Plotting transformed spectral data
‘ggspectra’ 0.3.17.9000
Pedro J. Aphalo
2026-02-20
Source:vignettes/articles/data-manipulation.Rmd
data-manipulation.RmdIntroduction
This article is only available as part of the on-line documentation of R package ‘ggspectra’. It is not installed in the local computer as part of the package.
Package ggspectra extends ggplot2 with
stats, geoms, scales and annotations suitable for light spectra. It also
defines ggplot() and autoplot() methods
specialized for the classes defined in package photobiology
for storing different types of spectral data. The
autoplot() methods are described separately in vignette
User Guide: 2 Autoplot Methods and the ggplot()
methods, statistics, and scales in User Guide: 1 Grammar of
Graphics.
The new elements can be freely combined with methods and functions
defined in packages ‘ggplot2’, scales,
ggrepel, gganimate, cowplot and
other extensions to ‘ggplot2’.
This article, formerly the third part of the User Guide, describes how to combine manipulation of spectral data with plotting. This streamlined coding is made possible by an enhancement implemented in ‘ggspectra’ (>= 0.3.5). In addition, some of the examples make use of methods available only in ‘photobiology’ (>= 0.10.0).
In ‘ggspectra’ (>= 0.3.5) the data member of gg
(ggplot) objects remains as an object of the classes for spectral data
defined in ‘photobiology’ instead of being converted into a plain
data.frame. This makes it possible data manipulations in
layers to be done with methods specific to spectral data. In other
words, the data object used as default for individual plot
layers retains its original attributes, including its class. This makes
it possible to use methods applicable to the original object to modify
it for individual plot layers.
The examples in this vignette depend conditionally on packages ‘rlang’, ‘magrittr’, and ‘gganimate’. If these packages are not available when the article is built, the code chunks that require them are not evaluated.
Set up
## Loading required package: SunCalcMeeus## Documentation at https://docs.r4photobiology.info/
library(photobiologyWavebands)
library(ggspectra)
# if suggested packages are available
magrittr_installed <- requireNamespace("magrittr", quietly = TRUE)
rlang_installed <- requireNamespace("rlang", quietly = TRUE)
eval_chunks <- magrittr_installed && rlang_installed
if (eval_chunks) {
library(magrittr)
library(rlang)
} else {
message("Please, install packages 'rlang' and 'magrittr'.")
}##
## Attaching package: 'rlang'## The following object is masked from 'package:magrittr':
##
## set_names
eval_gganimate <- requireNamespace("gganimate", quietly = TRUE)
if (eval_gganimate) {
library(gganimate)
message("'gganimate' version ",
format(packageVersion('gganimate')),
" loaded and attached.")
}## 'gganimate' version 1.0.11 loaded and attached.We change the default theme.
Applying methods to spectral data in plot layers
A single spectrum
In ‘ggspectra’ (< 0.3.5) we had to pass to the data
parameter of layer functions always a data frame, or a transformation
based on a method with an specialization for data,frame. A
simple example passing spectral data to each layer function follows.
Here to be able to use a method defined for source_spct
objects like sun.spct we pass sun.spct as
argument to method smooth_spct().
ggplot() +
geom_line(data = sun.spct, mapping = aes(w.length, s.e.irrad)) +
geom_line(data = smooth_spct(sun.spct, method = "supsmu"),
mapping = aes(w.length, s.e.irrad),
colour = "red", linewidth = 1)
In ‘ggspectra’ (< 0.3.5) we could also use R’s own pipe operator
to make it easier to see the intention of the code, but still had to
supply sun.spct twice.
ggplot() +
geom_line(data = sun.spct, mapping = aes(w.length, s.e.irrad)) +
geom_line(data = sun.spct |> smooth_spct(method = "supsmu"),
mapping = aes(w.length, s.e.irrad),
colour = "red", linewidth = 1)
In ‘ggspectra’ (>= 0.3.5) the class of the spectral objects stored
by calls to ggplot() methods specific to them is not
stripped, neither are other attributes used by package ‘photobiology’.
Consequently, transformations in layers using default data
can use the specialized methods from package ‘photobiology’. The next
two code chunks query the class and print a summary to demonstrate
this.
## [1] "source_spct" "generic_spct" "tbl_df" "tbl" "data.frame"
summary(p$data)## Summary of source_spct [522 x 2] object: anonymous
## Wavelength range 280-800 nm, step 0.9230769-1 nm
## Label: sunlight, simulated
## Measured on 2010-06-22 09:51:00 UTC
## Measured at 60.20911 N, 24.96474 E; Kumpula, Helsinki, FI
## Variables:
## w.length: Wavelength [nm]
## s.e.irrad: Spectral energy irradiance [W m-2 nm-1]
## --
## w.length s.e.irrad
## Min. :280.0 Min. :0.0000
## 1st Qu.:409.2 1st Qu.:0.4115
## Median :539.5 Median :0.5799
## Mean :539.5 Mean :0.5160
## 3rd Qu.:669.8 3rd Qu.:0.6664
## Max. :800.0 Max. :0.8205To ensure that the same data are used in both plot layers the code
can be simplified using . to refer to the default data in
the ggplot object (p$data in the example above). The
mapping to aesthetics remains valid because smooth_spct()
returns a new source_spct object with the same column names
as sun.spct, which was passed as argument to
data.
ggplot(sun.spct) +
geom_line() +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red", linewidth = 1)
Alternatively, without depending on ‘magrittr’ or ‘rlang’ we can pass
an anonymous function as argument to data to the same
effect, but possibly less clear code.
ggplot(sun.spct) +
geom_line() +
geom_line(data = function(x) {smooth_spct(x, method = "supsmu")},
colour = "red", linewidth = 1)
The easiest approach to plotting photon spectral irradiance instead
of spectral energy irradiance is to temporarily change the default
radiation unit. An alternative approach is to replace the first two
lines in the code chunk below by:
ggplot(sun.spct, unit.out = "photon") +.
photon_as_default()
ggplot(sun.spct) +
geom_line() +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red", linewidth = 1)
unset_radiation_unit_default()
Obviously, the default plot data does not need to be plotted, so this provides a roundabout way of applying methods,
ggplot(sun.spct) +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red", linewidth = 1)which is equivalent to doing the transformation ahead of plotting.
sun.spct |>
smooth_spct(method = "supsmu") |>
ggplot() +
geom_line(colour = "red", linewidth = 1)
However, when using different transformations in different layers we need to apply them in each layer. Here we compare three different smoothing methods.
ggplot(sun.spct) +
geom_line(data = . %>% smooth_spct(method = "custom"),
colour = "cornflowerblue", linewidth = 0.7) +
geom_line(data = . %>% smooth_spct(method = "lowess"),
colour = "green", linewidth = 0.7) +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red", linewidth = 0.7)
Of course, this approach works both with geoms and
stats, but one should remember that these layer functions do
not “see” the original data objects (neither the default nor that passed
as argument to the layers’ data parameter), but instead a
new data.frame containing the mapped variables in columns
named according to aesthetics. The next example demonstrates
this and illustrates that smoothing displaces the wavelength of maximum
spectral irradiance.
ggplot(sun.spct) +
geom_line() +
stat_peaks(size = 3, span = NULL) +
stat_peaks(geom = "vline", linetype = "dotted", span = NULL) +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red", linewidth = 1) +
stat_peaks(data = . %>% smooth_spct(method = "supsmu"),
colour = "red", size = 3, span = NULL) +
stat_peaks(data = . %>% smooth_spct(method = "supsmu"),
geom = "vline", colour = "red",
linetype = "dotted", span = NULL)
We can easily highlight a wavelength range by over-plotting the same line in a different colour.

We can highlight a range of wavelengths by plotting the points using
colours matching human colour vision. Method tag() adds
colour definitions as a new column named wl.color to the
default data of the layer, after wavelengths outside the
range of wavelengths of visible light have been dropped or “trimmed out”
by method trim_wl().
ggplot(sun.spct) +
geom_line() +
geom_point(data = . %>% trim_wl(range = VIS()) %>% tag(),
mapping = aes(color = wl.color),
shape = "circle", size = 1.3) +
scale_color_identity()
In the plot above, spectral irradiance as well as the wavelength at
each data point are taken into account when computing the colours, while
below, only the wavelength at the centre of each waveband is used
because we passed w.band = VIS_bands() when calling
tag().
ggplot(sun.spct) +
geom_area(data = . %>% trim_wl(range = VIS()) %>% tag(w.band = VIS_bands()),
mapping = aes(fill = wb.color)) +
geom_line() +
scale_fill_identity()
The examples above made use of sun.spct an object
belonging to class "source_spct", containing data for a
single spectrum. Package ‘photobiology’ defines classes for different
types of spectral data, and a base class "generic_spct".
Any object of belonging to one of these classes can be used as shown
above for sun.spct. Next we show examples involving
operations involving multiple spectra stored in a single R object.
Some of the methods from ‘photobiology’ are also defined for
data.frame and can be used as summary functions with data
that are not radiation spectra, such as any data
seen by layer functions including stat_summary().
Furthermore, on-the-fly summaries and transformation can be used in any
ggplot layer function and with any suitable function accepting data
frames as input.
Multiple spectra
Package ‘photobiology’ supports two alternative ways of storing
multiple spectra in a single R object: collections of spectra in objects
of classes with names ending in _mspct such as
source_mspct and in objects of classes with names ending in
_spct such as source_spct in objects. When
passed to ggplot() they are both added to the
"gg" object as objects of one of the _spct
classes. (The former are an specialization of lists of data frames,
while the second are an specialization of data frame, and, thus,
compatible with ‘ggplot2’.)
We use for these examples data for a time series of five spectra
measured close to sunset. Objects sun_evening.spct and
sun_evening.mspct contain the same data but belong to
classes "source_spct" and "source_mspct",
respectively.
We use the two spectra extracted from the collection
sun_evening.mspct, as two spectra are enough to demonstrate
how on-the-fly operations work with multiple spectra. This first example
is just a plain plot with two panels.
ggplot(data = sun_evening.mspct[c(2, 5)]) +
aes(linetype = spct.idx) +
geom_line() +
facet_wrap(facets = vars(spct.idx), ncol = 1)
Similarly as shown above for a single spectrum, we can smooth multiple spectra.
ggplot(data = sun_evening.mspct[c(2, 5)]) +
aes(linetype = spct.idx) +
geom_line(colour = "blue", alpha = 0.5) +
geom_line(data = . %>% smooth_spct(method = "supsmu")) +
facet_wrap(facets = vars(spct.idx), ncol = 1)
Normalizing the spectra to one at their maxima gives a clearer view of how they differ in shape. However, an annotation of the plot giving the irradiance before normalization can help understand the data.
ggplot(data = sun_evening.mspct[c(2, 5)]) +
aes(linetype = spct.idx) +
geom_line(data = . %>% normalize()) +
stat_wb_irrad(time.unit = "second",
unit.in = "energy",
w.band = list(PAR(), NIR()),
label.fmt = "%.1#f~W~m^{-2}",
parse = TRUE,
colour = "black",
size = 2.5) +
facet_wrap(facets = vars(spct.idx), ncol = 1)
It can be even clearer to scale the spectra so that the areas under the different curves are the same.
ggplot(data = sun_evening.mspct[c(2, 5)]) +
aes(linetype = spct.idx) +
geom_line(data = . %>% fscale()) +
stat_wb_irrad(time.unit = "second",
unit.in = "energy",
w.band = list(PAR(), NIR()),
label.fmt = "%.1#f~W~m^{-2}",
parse = TRUE,
colour = "black",
size = 2.5) +
facet_wrap(facets = vars(spct.idx), ncol = 1)
Transforming data when calling layer functions is
useful, as shown here, when different transformations to the data are
applied in different layers of the same plot.
While above we used two spectra extracted from
sun_evening.mspct, here we use
sun_evening.spct, and plot all five spectra.
ggplot(data = sun_evening.spct) +
aes(linetype = spct.idx) +
geom_line() +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red") +
facet_wrap(facets = vars(spct.idx), ncol = 2)
Another use case is when a layer displays a summary spectrum computed
from multiple spectra shown in a different plot layer. In the example
below the blue line shows the median spectrum. Other spectra are
semitransparent and show as grey lines. To avoid the mapping to
group for the plot as a whole we pass aes() as
mapping to be merged with the automatic mapping of x and
y.
ggplot(data = sun_evening.spct, aes()) +
geom_line(aes(group = .data[[id_factor(sun_evening.spct)]]),
alpha = 1/3) +
geom_line(data = . %>% s_median(), # parallel median
colour = "blue")
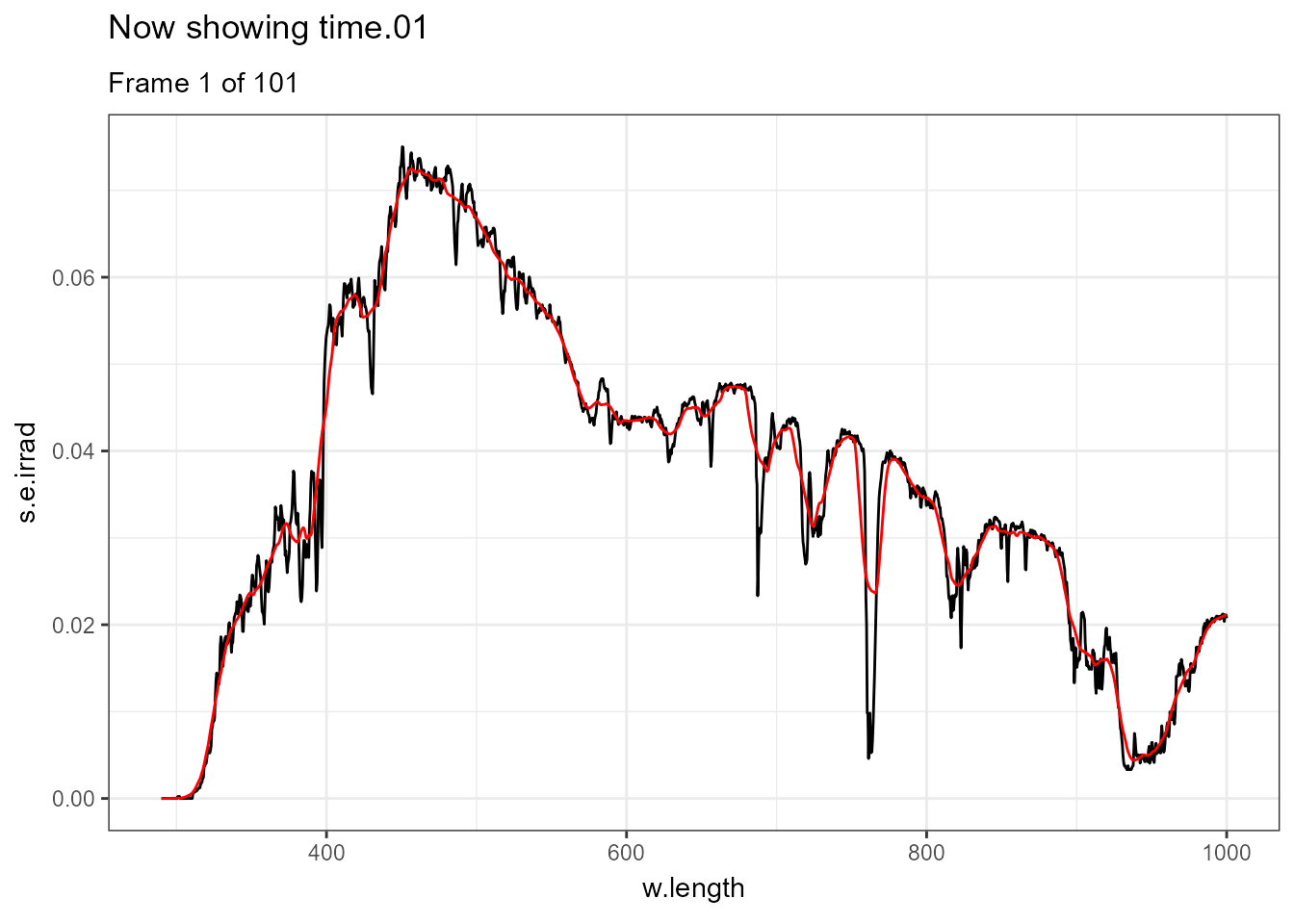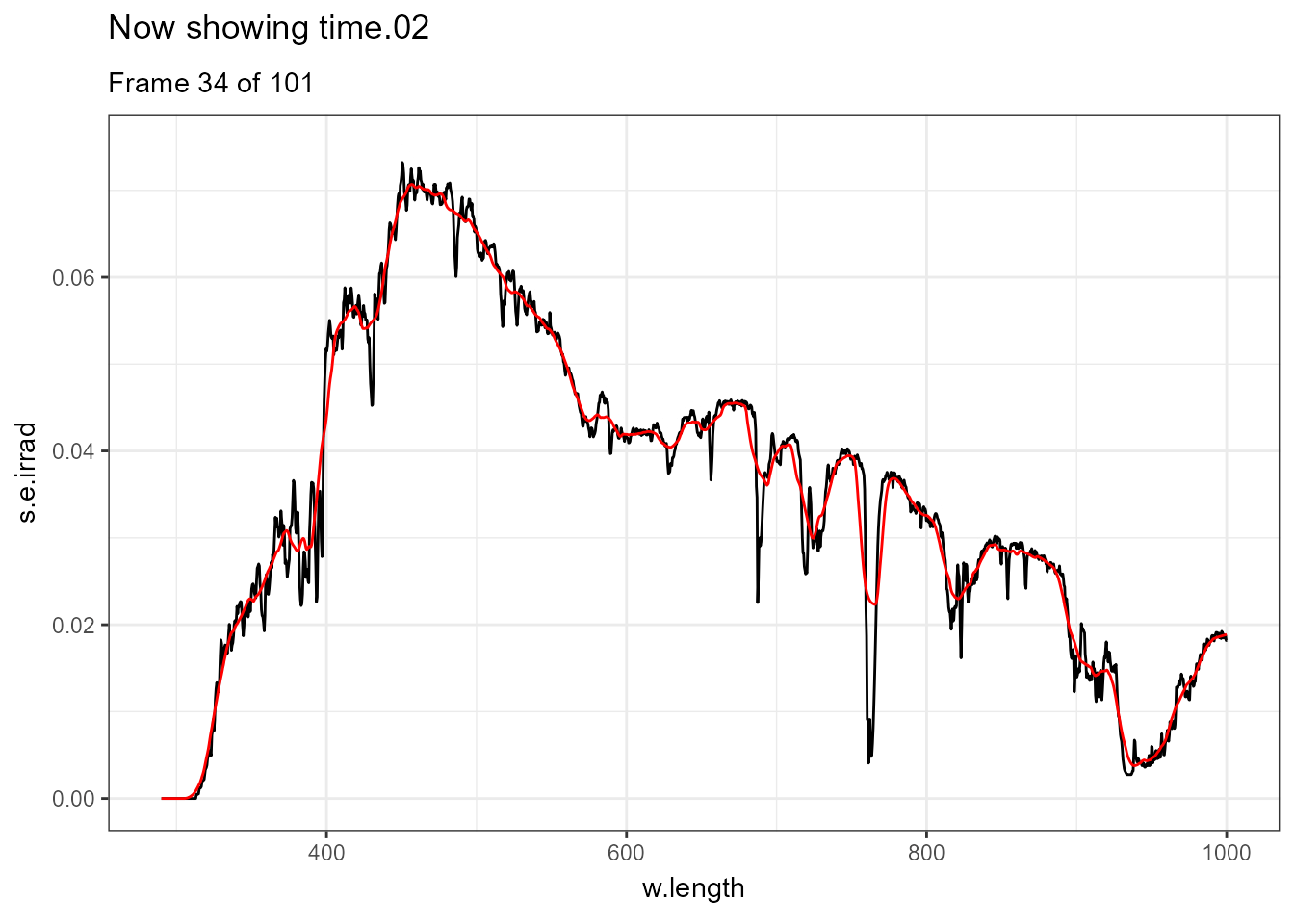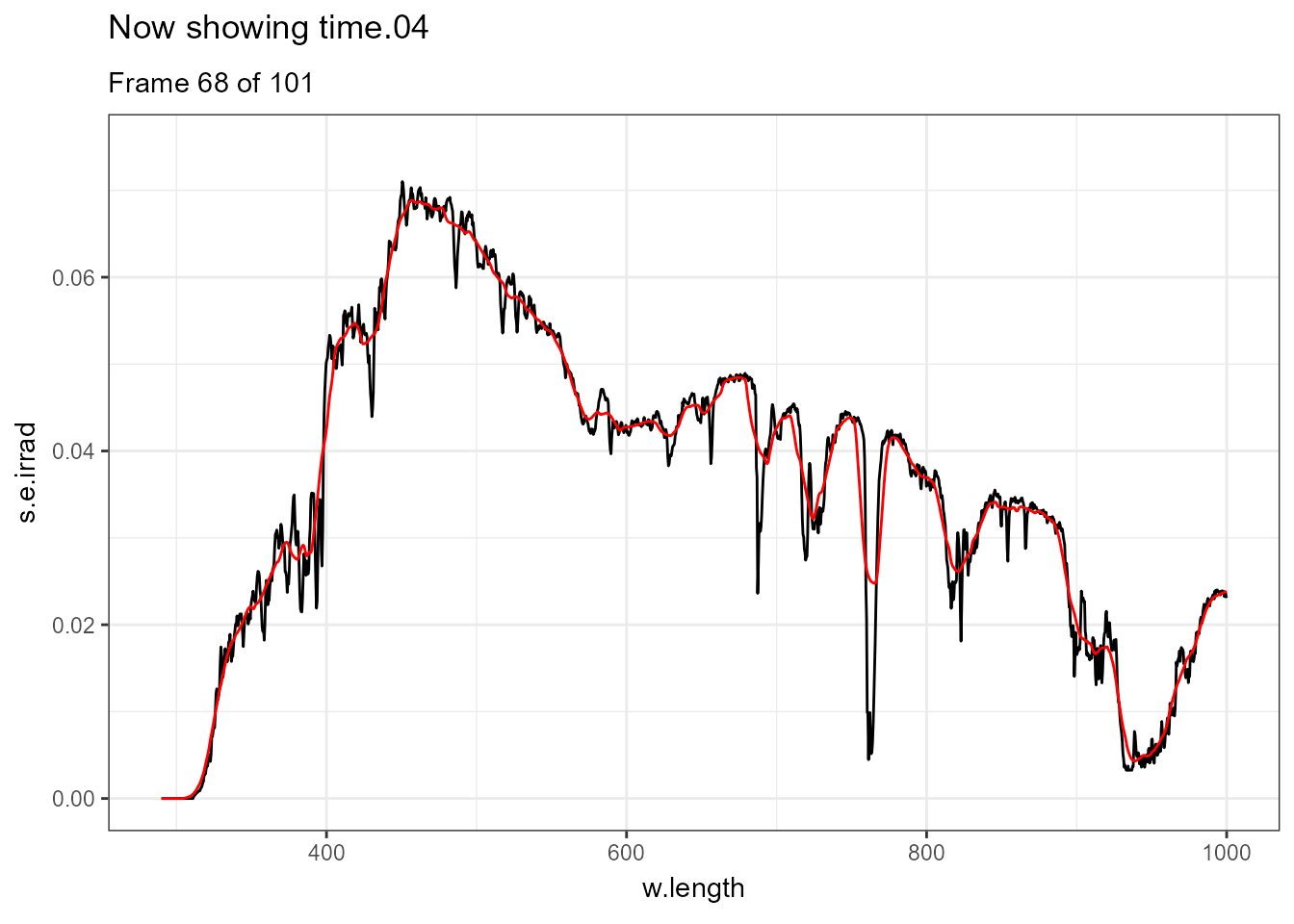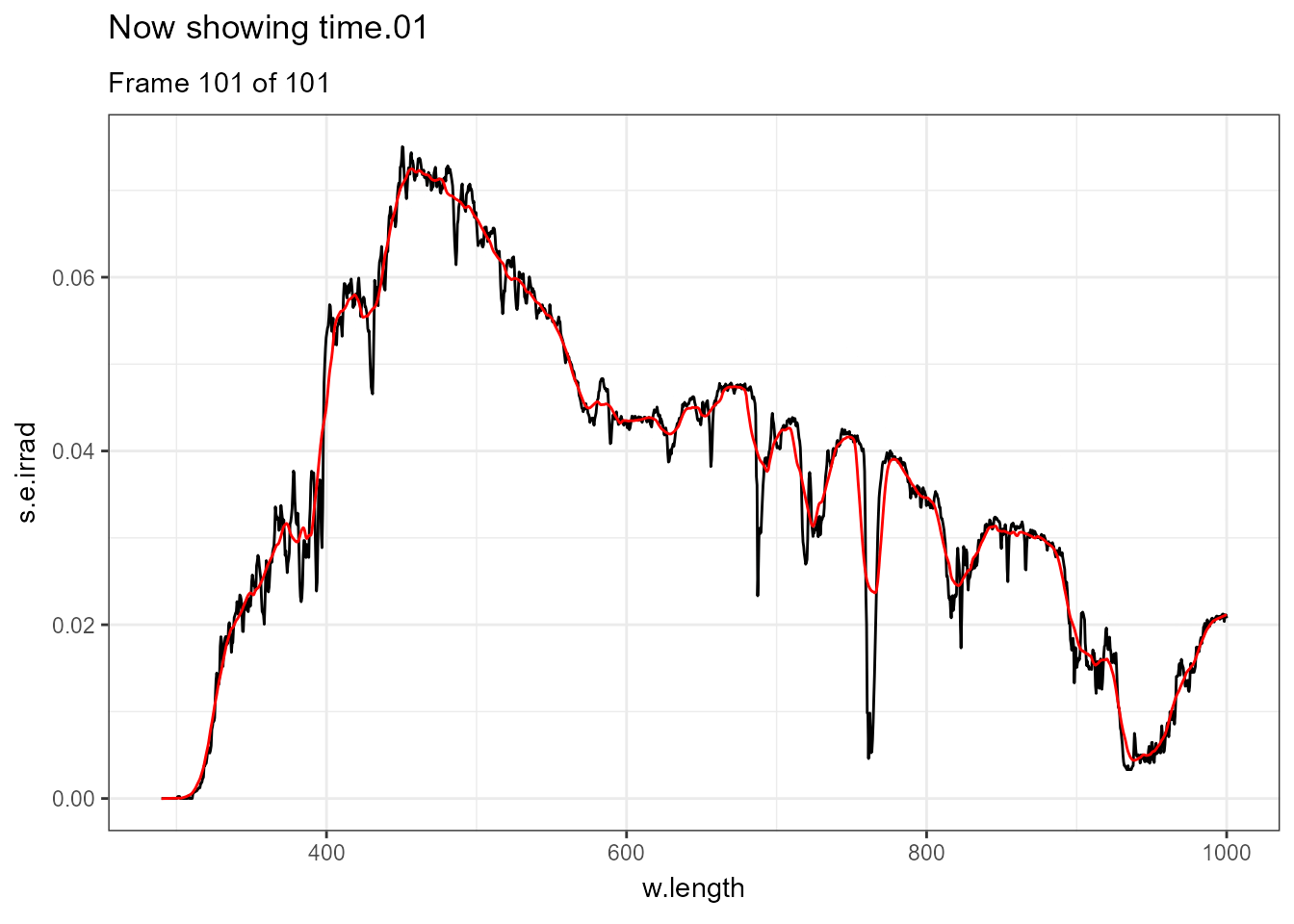
There are few situations where applying transformations to multiple spectra does not result in overcrowded plots, one example being animated plots where different layers are displayed sequentially. Further examples of animations are available in the separate on-line only package article Animated plots of spectral data. In this case a smoothed version of each spectrum is displayed in red and the measured spectrum in black.
ggplot(data = sun_evening.mspct) +
geom_line() +
geom_line(data = . %>% smooth_spct(method = "supsmu"),
colour = "red") +
transition_states(spct.idx,
transition_length = 2,
state_length = 1) +
ggtitle('Now showing {closest_state}',
subtitle = 'Frame {frame} of {nframes}')