
User Guide 1: The data and how to use them
Package ‘photobiologyFilters’ 0.6.1
Pedro J. Aphalo
2025-09-23
Source:vignettes/user-guide.Rmd
user-guide.RmdIntroduction
This package, is a data only package, part of a suite, which has package ‘photobiology’ at its core. Please visit (https://www.r4photobiology.info/) for additional information. For details on plotting spectra, please consult the documentation for package ‘ggspectra’, and for information on the calculation of summaries and maths operations between spectra, please, consult the documentation for package ‘photobiology’.
## Loading required package: SunCalcMeeus## Documentation at https://docs.r4photobiology.info/## Loading required package: ggplot2In this brief User Guide we describe how to access individual spectra or subsets of spectra.
The objects containing the data
The data are stored in objects of classes defined in package ‘photobiology’ grouped into four collections of spectra. Functions and operators in the packages that are part of the R for photobiology suite recognize these classes, which allows automation of some computations and validation of the data values returned by operations on the data.
Individual spectra are stored in clases are derived from tibbles,
which in turn is derived from data frames with additional metadata
stored as attributes of the objects. Being the objects derived from
data.frame means that the data can be used with native R
functions and operators and with other R packages. As package
‘photobiologyInOut’ provides functions that facilitate the exchange of
spectral data with R packages ‘colorSpec’, ‘hyperSpec’ and ‘pavo’, the
data in these package can also be easily used with them.
The individual spectra are stored in collections as objects of classes derived from R’s list class.
filters.mspct contains spectral transmittance data for
389 optical filters and other similar transparent and semitransparent
sheets and films.
class(filters.mspct)## [1] "filter_mspct" "generic_mspct" "list"
class(filters.mspct[[1]])## [1] "filter_spct" "generic_spct" "tbl_df" "tbl" "data.frame"filters_UVIVIF.mspct contains UV-A1-induced spectral
fluorescence data for 7 optical filters.
class(filters_UVIVIF.mspct)## [1] "source_mspct" "generic_mspct" "list"
class(filters_UVIVIF.mspct[[1]])## [1] "source_spct" "generic_spct" "tbl_df" "tbl" "data.frame"metals.mspct contains spectral reflectance data for 12
metals.
class(metals.mspct)## [1] "reflector_mspct" "generic_mspct" "list"
class(metals.mspct[[1]])## [1] "reflector_spct" "generic_spct" "tbl_df" "tbl"
## [5] "data.frame"materials.mspct contains spectral reflectance data for
12 materials and surfaces, including different types vegetation.
class(materials.mspct)## [1] "reflector_mspct" "generic_mspct" "list"
class(materials.mspct[[1]])## [1] "reflector_spct" "generic_spct" "tbl_df" "tbl"
## [5] "data.frame"refractive_index.mspct contains spectral refractive
index data for 12 materials. These values allow the computation of
reflectance at different angles of incidence.
class(refractive_index.mspct)## [1] "generic_mspct" "list"
class(refractive_index.mspct[[1]])## [1] "generic_spct" "tbl_df" "tbl" "data.frame"Spectral reflectance and transmittance values are stored as fractions of one, and wavelengths expressed in nanometres (nm).
colnames(filters.mspct[[1]])## [1] "w.length" "Tfr"
colnames(metals.mspct[[1]])## [1] "w.length" "Rfr"
colnames(materials.mspct[[1]])## [1] "w.length" "Rfr"Refractive index values are:
colnames(refractive_index.mspct[[1]])## [1] "w.length" "n" "k"The transmittance and reflectance data
Spectral data are included both for optical filters, sold as such, and for materials that either on purpose or by accident may be interposed in the “light” path in photobiological experiments, including glass panes, plastic sheets and films. It must be kept in mind that, 1) absorptance depends on the thickness of a filter in addition to the properties of the material it is made off, and that 2) reflectance depends on the angle of incidence of the light beam. Spectral transmittance can be expressed either as internal or total, depending on how the effect reflections at surfaces are accounted for. The conversion between total and internal transmittance requires reflectance to be known.
We provide reflectance estimates for several of the filters in this
collection, but we do not have such data available for all of them. For
filters made of ionic glass and coloured plastics, reflection is very
weakly selective, and amounts to about 9 to 10% of radiation incident at
an angle close to perpendicular to the surface (when both interfaces
with air are considered). Anti-reflection coating (ARC) reduces
reflections, and multi-coating (MC) even further. Such coatings are not
equally effective at all wavelengths and, consequently, their use can
modify the spectral properties of a filter. In contrast to absorptive
filters described above, dichroic or interference filters reflect the
“rejected” radiation. It is also possible to produce filters that have
an absorbing glass as substrate and a dichroic coating deposited onto
one or both of its surfaces. In the metadata filters are tagged as
belonging to one of three attenuation.mode types:
absorption, reflection and mixed.
Mixed includes those filters where wavelength-selective attenuation is
brought about both by absorption and reflection, including scattering
media and dichroic filters deposited on absorptive ionic glass.
The spectral transmittance or absorbance data have been acquired with an assortment of different instruments. Some data are measured by the authors with spectrophotometers, others have been provided by filter manufacturers. Whenever these metadata were available an approximate reflectance for normal incidence angle, material thickness, and the mode of attenuation have been stored as attributes. In the case of filters sold assembled in frames for use in photography and industrial automation the thickness is rarely disclosed by suppliers, but in some cases we have been able to measure it.
For absortive filters with no coatings reflectance was computed from the spectral refraction index if available, or copied from manufacturer’s specifications or for some standard materials based on published sources. In the case of filters with anti-reflection coatings, in most cases the estimated reflectance is based on the maximum total transmittance. Most of the data are approximations given as a single value for the whole spectrum following Schott’s practice. (We aim to add in the future actual measured and/or computed spectral reflectance for some filters or clear glass to serve as examples).
The difference in resolution and slit function among instruments can give, for the same filter, measured spectra with “apparent” peaks and valleys of slightly different width, and slopes of slightly different steepness. This is an inevitable artifact of spectral measurements, which has little effect on the spectra from absorptive filters (with gradual slopes and bell-shaped peaks) but can be more significant for dichroic (= interference) filters with very steep cut-on and cut-off slopes.
Another important consideration is that some materials scatter transmitted and reflected light, and consequently such materials can be accurately measured only with an integrating sphere. Data included here have been in many cases measured without an integrating sphere; i.e., only by assessing the direct beam. For non-scattering materials this causes only minor errors. For scattering materials like polythene films, the data included have being measured with an integrating sphere.
Glass-filter properties vary to some extent among melt batches. Variation can also be expected among batches of plastic filters. Furthermore, filters age upon exposure to light and UV radiation, and in some cases even upon exposure to air. Aging is not limited to plastic filters and can also affect optical glass. Manufacturers occasionally may update the specifications for filters while maintaining the product code.
All the spectral transmittance data in this package are stored in a
single R object, a collection of spectra of class
filter_mspct with members of class
filter_spct. Individual or subsets of spectra can be
retrieved by name. The package includes also several
character vectors of names, each one containing
names for filters of a given color, a given type or
from a given manufacturer. The names of all these vectors are
available in vector all_filter_selectors. The
names used are in most cases the codes used by the
manufacturers for the given type with a code for the manufacturer of
supplier prepended. Any dashes in these codes have been replaced by
underscores to convert them into syntactically valid R names.
band_pass_filters## [1] "BPI_Luminance" "BPI_Solatrol"
## [3] "Foiltek_Clear_PET_G" "Heliopan_BG38_2.3mm_46mm"
## [5] "McDermit_PET_Autostat_CT5_125um" "MIDOPT_AB555"
## [7] "MIDOPT_Bi405" "MIDOPT_Bi440"
## [9] "MIDOPT_Bi450" "MIDOPT_Bi518"
## [11] "MIDOPT_Bi520" "MIDOPT_Bi550"
## [13] "MIDOPT_Bi615" "MIDOPT_Bi632"
## [15] "MIDOPT_Bi650" "MIDOPT_Bi660"
## [17] "MIDOPT_Bi685" "MIDOPT_Bi725"
## [19] "MIDOPT_Bi780" "MIDOPT_Bi808"
## [21] "MIDOPT_Bi830" "MIDOPT_Bi850"
## [23] "MIDOPT_Bi880" "MIDOPT_Bi905"
## [25] "MIDOPT_BN470" "MIDOPT_BN485"
## [27] "MIDOPT_BN490" "MIDOPT_BN532"
## [29] "MIDOPT_BN535" "MIDOPT_BN595"
## [31] "MIDOPT_BN630" "MIDOPT_BN650"
## [33] "MIDOPT_BN660" "MIDOPT_BN740"
## [35] "MIDOPT_BN785" "MIDOPT_BN810"
## [37] "MIDOPT_BN840" "MIDOPT_BN850"
## [39] "MIDOPT_BN880" "MIDOPT_BN940"
## [41] "MIDOPT_BP250" "MIDOPT_BP324"
## [43] "MIDOPT_BP365" "MIDOPT_BP470"
## [45] "MIDOPT_BP485" "MIDOPT_BP500"
## [47] "MIDOPT_BP505" "MIDOPT_BP525"
## [49] "MIDOPT_BP550" "MIDOPT_BP590"
## [51] "MIDOPT_BP635" "MIDOPT_BP660"
## [53] "MIDOPT_BP695" "MIDOPT_BP735"
## [55] "MIDOPT_BP800" "MIDOPT_BP810"
## [57] "MIDOPT_BP845" "MIDOPT_BP850"
## [59] "MIDOPT_BP865" "MIDOPT_BP880"
## [61] "MIDOPT_DB395_870" "MIDOPT_DB475_850"
## [63] "MIDOPT_DB550_850" "MIDOPT_DB660_850"
## [65] "MIDOPT_DB735" "MIDOPT_DB850"
## [67] "MIDOPT_DB940" "MIDOPT_PE530"
## [69] "MIDOPT_TB475_550_850" "MIDOPT_TB550_660_850"
## [71] "Schott_BG18" "Schott_BG25"
## [73] "Schott_BG3" "Schott_BG36"
## [75] "Schott_BG38" "Schott_BG39"
## [77] "Schott_BG40" "Schott_BG42"
## [79] "Schott_BG50" "Schott_BG55"
## [81] "Schott_BG60" "Schott_BG60HT"
## [83] "Schott_BG61" "Schott_BG62"
## [85] "Schott_BG62HS" "Schott_BG62HT"
## [87] "Schott_BG63" "Schott_BG64"
## [89] "Schott_BG66HS" "Schott_BG67"
## [91] "Schott_BG7" "Schott_UG1"
## [93] "Schott_UG11" "Schott_UG2A"
## [95] "Schott_UG5" "Unbranded_Clear_LD_PE_50um"
## [97] "Unbranded_Clear_LD_PE_50um_used" "UQG_Blue_dichroic_CDB"
## [99] "UQG_UG1_2mm"
schott_filters## [1] "Schott_UG1" "Schott_UG5" "Schott_UG11" "Schott_BG3"
## [5] "Schott_BG7" "Schott_BG18" "Schott_BG25" "Schott_BG36"
## [9] "Schott_BG38" "Schott_BG39" "Schott_BG40" "Schott_BG42"
## [13] "Schott_S_8612" "Schott_BG50" "Schott_BG55" "Schott_BG60"
## [17] "Schott_BG60HT" "Schott_BG61" "Schott_BG62" "Schott_BG62HT"
## [21] "Schott_BG63" "Schott_BG64" "Schott_VG9" "Schott_VG20"
## [25] "Schott_S_8022" "Schott_S_8023" "Schott_GG395" "Schott_GG400"
## [29] "Schott_GG420" "Schott_GG435" "Schott_GG455" "Schott_GG475"
## [33] "Schott_GG495" "Schott_OG515" "Schott_OG530" "Schott_OG550"
## [37] "Schott_OG570" "Schott_OG590" "Schott_RG9" "Schott_RG610"
## [41] "Schott_RG630" "Schott_RG645" "Schott_RG665" "Schott_RG695"
## [45] "Schott_RG715" "Schott_RG780" "Schott_RG830" "Schott_RG850"
## [49] "Schott_RG1000" "Schott_NG1" "Schott_NG3" "Schott_NG4"
## [53] "Schott_NG5" "Schott_NG9" "Schott_NG11" "Schott_N_WG280"
## [57] "Schott_N_WG295" "Schott_N_WG305" "Schott_N_WG320" "Schott_KG1"
## [61] "Schott_KG2" "Schott_KG3" "Schott_KG5" "Schott_BG67"
## [65] "Schott_UG2A" "Schott_BG62HS" "Schott_BG66HS"Help pages give additional information.
help(schott_filters)In addition to these data for filters, the package contains spectral reflectance data for both metal surfaces and various man-made and natural surfaces. All spectral reflectance data in the package are for normally incident radiation and total values, that is including both especular reflection and scattering.
These collections are small, so we list the names of all members.
names(metals.mspct)## [1] "aluminum" "copper" "gold" "molybdenum"
## [5] "nickel" "platinum" "silver" "stainless_steel"
## [9] "tantalum" "tin" "titanium" "vanadium"
names(materials.mspct)## [1] "construction.asphalt" "construction.concrete."
## [3] "aluminum.metal" "galvanized.steel.metal"
## [5] "light.yellowish.brown.clay" "black.loam"
## [7] "dry.grass" "grass"
## [9] "conifer" "decidous"
## [11] "fine.snow" "coarse.granular.snow"
## [13] "medium.granular.snow" "gold.plate"
names(refractive_index.mspct)## [1] "Ag" "air" "Al" "Au" "Cr" "Cu" "Ge" "NBAF10"
## [9] "NBK7" "Ni" "PC" "PET" "PMMA" "PS" "PVC" "Ti"
## [17] "water"The collection of filter spectra is large, so we list only the first 20 names.
## [1] "Baader_U_filter_1.0mm_48mm" "BPI_Luminance"
## [3] "BPI_Solatrol" "BW_007_Clear_MRC_nano_1.2mm_46mm"
## [5] "Courtaulds_CA_115um" "Courtaulds_CA_115um_age000"
## [7] "Courtaulds_CA_115um_age020" "Courtaulds_CA_115um_age030"
## [9] "Courtaulds_CA_115um_age060" "Courtaulds_CA_115um_age100"
## [11] "Courtaulds_CA_115um_age180" "Courtaulds_CA_115um_age300"
## [13] "Courtaulds_CA_250um" "Courtaulds_CA_480um"
## [15] "Evonik_Cherry_3C01_GT" "Evonik_Clear_0A000_XT"
## [17] "Evonik_Clear_0F00_GT" "Evonik_Clear_2458_GT"
## [19] "Evonik_Clear_UV_100_0A570_GT" "Evonik_Orange_2C04_GT"To obtain metadata in a tabular form, we can use function
spct_metadata(). This function is available in package
‘photobiology’ version 0.10.5 or later. (This example is run only if a
supported version is loaded.) The limit the number of columns we select
two of them by name.
if (utils::packageVersion('photobiology') > "0.10.4.9001") {
print(colnames(spct_metadata(filters.mspct[1:5])))
spct_metadata(filters.mspct[1:5])[ , c("spct.idx", "what.measured")]
}## [1] "spct.idx" "what.measured" "how.measured" "normalized"
## [5] "multiplier" "Tfr.type"## # A tibble: 5 × 2
## spct.idx what.measured
## <fct> <chr>
## 1 Baader_U_filter_1.0mm_48mm Astrophotography filter: Baader U filter
## 2 BPI_Luminance Luminance, special greenhouse cladding film …
## 3 BPI_Solatrol Solatrol, special greenhouse cladding film f…
## 4 BW_007_Clear_MRC_nano_1.2mm_46mm Photography filter: BW 007 Clear MRC nano
## 5 Courtaulds_CA_115um Courtaulds cellulose (di-)acetate (CA); 0.00…Accessing individual spectra
The filter_spct member objects in
filters.mspct and the reflector_spct objects
contained in metals.mspct and materials.mspct
can be accessed through their names or through a numeric index. As the
numeric indexes are likely to change with updates to the package, their
use is discouraged. Names should be used instead. The names are listed
in the documentation and be listed with method names(). All
examples below use filters.mspct but operations on the
other collections work in the same way.
We can use a character string with the name as an index to extract an individual spectrum.
filters.mspct$Schott_UG11## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00001
## 2 204 0.00001
## 3 207 0.00001
## 4 208 0.00001
## 5 209 0.00001
## 6 210 0.00001
## 7 211 0.00001
## 8 212 0.00001
## 9 213 0.00001
## 10 214 0.00001
## # ℹ 696 more rows
filters.mspct[["Schott_UG11"]]## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00001
## 2 204 0.00001
## 3 207 0.00001
## 4 208 0.00001
## 5 209 0.00001
## 6 210 0.00001
## 7 211 0.00001
## 8 212 0.00001
## 9 213 0.00001
## 10 214 0.00001
## # ℹ 696 more rowsBe aware that according to R’s rules, using single square brackets
will return a filter_mspct object possibly of length one.
This statement is not equivalent to the one in the chunk immediately
above.
filters.mspct["Schott_UG11"]## Object: filter_mspct [1 x 1]
## --- Member: Schott_UG11 ---
## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00001
## 2 204 0.00001
## 3 207 0.00001
## 4 208 0.00001
## 5 209 0.00001
## 6 210 0.00001
## 7 211 0.00001
## 8 212 0.00001
## 9 213 0.00001
## 10 214 0.00001
## # ℹ 696 more rows
##
## --- END ---Of course, with single square brackets it is possible to use a vector of member names.
Accessing subsets of spectra
We can subset the filter_mspct object by indexing with
vectors of character strings. The package provides several predefined
ones, and users can easily define their own, either as constants or
through computation. Here we use a vector defined by the package.
filters.mspct[petri_dishes]## Object: filter_mspct [3 x 1]
## --- Member: glass_nn ---
## Object: filter_spct [325 x 2]
## Wavelength range 190-1100 nm, step 1-4 nm
## Label: Petri dish lid; glass
## Rfr (/1): 0.080, thickness (mm): NA, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Total spectral transmittance [/1]
## --
## # A tibble: 325 × 2
## w.length Tfr
## <int> <dbl>
## 1 190 0.000142
## 2 194 0.000129
## 3 198 0.000117
## 4 202 0.000105
## 5 206 0.000102
## 6 207 0.000102
## 7 210 0.000103
## 8 214 0.000106
## 9 218 0.000106
## 10 219 0.000106
## # ℹ 315 more rows
## --- Member: PS_Sterilin101 ---
## Object: filter_spct [333 x 2]
## Wavelength range 190-1100 nm, step 1-4 nm
## Label: Petri dish lid; polystyrene; Sterilin101
## Rfr (/1): 0.104, thickness (mm): NA, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Total spectral transmittance [/1]
## --
## # A tibble: 333 × 2
## w.length Tfr
## <int> <dbl>
## 1 190 0.000127
## 2 194 0.000119
## 3 198 0.000111
## 4 202 0.000105
## 5 206 0.000100
## 6 207 0.000100
## 7 208 0.000100
## 8 210 0.000100
## 9 214 0.000106
## 10 218 0.000108
## # ℹ 323 more rows
## --- Member: PS_Sterilin109 ---
## Object: filter_spct [318 x 2]
## Wavelength range 190-1100 nm, step 1-4 nm
## Label: Petri dish lid; polystyrene; Sterilin109
## Rfr (/1): 0.098, thickness (mm): NA, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Total spectral transmittance [/1]
## --
## # A tibble: 318 × 2
## w.length Tfr
## <int> <dbl>
## 1 190 0.000134
## 2 194 0.000124
## 3 198 0.000115
## 4 202 0.000104
## 5 206 0.000106
## 6 210 0.000108
## 7 213 0.000112
## 8 214 0.000112
## 9 218 0.000112
## 10 222 0.000107
## # ℹ 308 more rows
##
## --- END ---The vector all_filter_selectors contains the names of
the different vectors of names of members of
filters.mspct.
all_filter_selectors## [1] "acetate_filters" "acrylic_filters"
## [3] "baader_filters" "band_pass_filters"
## [5] "blue_filters" "blue_green_filters"
## [7] "bpi_visqueen_filters" "bw_filters"
## [9] "clear_filters" "courtaulds_filters"
## [11] "etola_filters" "evonik_filters"
## [13] "fake_unbranded_filters" "firecrest_filters"
## [15] "foiltek_filters" "fotga_filters"
## [17] "glass_windows" "green_filters"
## [19] "haida_filters" "heat_filters"
## [21] "heliopan_filters" "hoya_filters"
## [23] "kenfaith_filters" "kenko_filters"
## [25] "knightx_filters" "kolarivision_filters"
## [27] "lee_filters" "lee_gels"
## [29] "long_pass_filters" "mcdermit_filters"
## [31] "midopt_filters" "neutral_filters"
## [33] "nisi_filters" "old_schott_filters"
## [35] "optical_glass_filters" "orange_filters"
## [37] "photography_filters" "plastic_dome_filters"
## [39] "plastic_film_filters" "plastic_sheet_filters"
## [41] "plexiglas_filters" "polycarbonate_filters"
## [43] "polyester_filters" "polystyrene_filters"
## [45] "polyvynil_chloride_filters" "purshee_filters"
## [47] "red_nir_filters" "rocolax_filters"
## [49] "rosco_filters" "rosco_gels"
## [51] "schott_filters" "short_pass_filters"
## [53] "stacked_filters" "tangsinuo_filters"
## [55] "theatrical_gels" "tiffen_filters"
## [57] "uqg_filters" "uv_filters"
## [59] "uvir_cut_filters" "uvroptics_filters"
## [61] "xl_horticulture_filters" "yellow_filters"
## [63] "zeiss_filters" "zomei_filters"In addition to the predefined vectors it is possible to compute
numeric indexing vectors using pattern matching with
grep(). In this example we extract the member spectra with
names containing the string “UG”.
## Object: filter_mspct [5 x 1]
## --- Member: Schott_UG1 ---
## Object: filter_spct [613 x 2]
## Wavelength range 258-5200 nm, step 1-50 nm
## Label: SCHOTT UG1 1mm-thick
## Rfr (/1): 0.086, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 613 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 258 4.30e-12
## 2 262 2.63e- 9
## 3 266 2.96e- 7
## 4 270 9.86e- 6
## 5 274 1.45e- 4
## 6 278 1.05e- 3
## 7 279 1.60e- 3
## 8 280 2.38e- 3
## 9 281 3.43e- 3
## 10 282 4.81e- 3
## # ℹ 603 more rows
## --- Member: Schott_UG11 ---
## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00001
## 2 204 0.00001
## 3 207 0.00001
## 4 208 0.00001
## 5 209 0.00001
## 6 210 0.00001
## 7 211 0.00001
## 8 212 0.00001
## 9 213 0.00001
## 10 214 0.00001
## # ℹ 696 more rows
## --- Member: Schott_UG2A ---
## Object: filter_spct [653 x 2]
## Wavelength range 269-5200 nm, step 1-55 nm
## Label: SCHOTT UG2A 3.25mm-thick
## Rfr (/1): 0.082, thickness (mm): 3.25, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 653 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 269 4.12e-22
## 2 273 1.54e-16
## 3 277 3.15e-12
## 4 281 4.69e- 9
## 5 285 8.80e- 7
## 6 289 3.98e- 5
## 7 293 6.29e- 4
## 8 294 1.09e- 3
## 9 295 1.84e- 3
## 10 296 2.95e- 3
## # ℹ 643 more rows
## --- Member: Schott_UG5 ---
## Object: filter_spct [652 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG5 1mm-thick
## Rfr (/1): 0.086, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 652 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00001
## 2 204 0.00001
## 3 207 0.00001
## 4 208 0.00001
## 5 209 0.00001
## 6 212 0.000315
## 7 213 0.000817
## 8 214 0.00200
## 9 215 0.00452
## 10 216 0.0109
## # ℹ 642 more rows
## --- Member: UQG_UG1_2mm ---
## Object: filter_spct [401 x 2]
## Wavelength range 190-1020 nm, step 1-4 nm
## Label: Glass filter: UQG UG1 2mm
## Rfr (/1): 0.077, thickness (mm): 2.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Total spectral transmittance [/1]
## --
## # A tibble: 401 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 190 0.000112
## 2 194 0.000107
## 3 198 0.000102
## 4 202 0.000102
## 5 206 0.000101
## 6 210 0.000100
## 7 214 0.000100
## 8 217 0.000100
## 9 218 0.000100
## 10 219 0.000100
## # ℹ 391 more rows
##
## --- END ---To generate the subset of names matching a pattern, we can also use
grep().
## [1] "Schott_UG1" "Schott_UG11" "Schott_UG2A" "Schott_UG5" "UQG_UG1_2mm"Querying metadata
The spectra are saved in objects of class "filter_spct",
defined in package ‘photobiology’. Specializations of several methods
including print() and summary() include a
summary of the metadata in the header of the printout. Two different
definitions of transmittance exist, differing in how reflection is
treated: for “internal” transmittance, the divisor is the radiation
entering the material, and for “total” transmittance the incident
radiation. For some materials reflectance (Rfr) does not
vary much with wavelength, and some suppliers provide a constant value
for it.
filters.mspct$Schott_UG11## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00001
## 2 204 0.00001
## 3 207 0.00001
## 4 208 0.00001
## 5 209 0.00001
## 6 210 0.00001
## 7 211 0.00001
## 8 212 0.00001
## 9 213 0.00001
## 10 214 0.00001
## # ℹ 696 more rowsMetadata can also be queried with other methods. Please, see the documentation for package ‘photobiology’ for the details.
filter_properties(filters.mspct$Schott_UG11)## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
what_measured(filters.mspct$Schott_UG11)## [1] "SCHOTT UG11 1mm-thick"
how_measured(filters.mspct$Schott_UG11)## [1] "Numerical data from supplier."
is_normalized(filters.mspct$Schott_UG11)## [1] FALSE## SCHOTT filter 'UG11' data, reference thickness (m): 0.001 and reflectance factor: 0.908
## (c) copyright SCHOTT, reproduced with permission.Of the different metadata items, the type of data is of great importance. Transmittance (and absorptance) can be expressed in two different ways: 1) taking incident radiation as reference, or 2) taking radiation entering the material as reference (i.e. discounting reflection). 1) is referred as total transmittance while 2) is referred as internal transmittance. Attribute `Tfr.type” is used to store this information.
getTfrType(filters.mspct$Schott_UG11)## [1] "internal"Conversions
When metatdata are available and the mode of attenuation is
"absorption" it is possible to compute the expected
transmittance for a different thickness of the material. In the example
we compute transmittance for a thickness of 4 mm.
convertThickness(filters.mspct$Schott_UG11, thickness = 4e-3)## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 4.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Internal spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 1e-20
## 2 204 1e-20
## 3 207 1e-20
## 4 208 1e-20
## 5 209 1e-20
## 6 210 1e-20
## 7 211 1e-20
## 8 212 1e-20
## 9 213 1e-20
## 10 214 1e-20
## # ℹ 696 more rowsWe can also convert "internal" transmittance into
"total" transmittance.
convertTfrType(filters.mspct$Schott_UG11, Tfr.type = "total")## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Tfr: Total spectral transmittance [/1]
## --
## # A tibble: 706 × 2
## w.length Tfr
## <dbl> <dbl>
## 1 200 0.00000908
## 2 204 0.00000908
## 3 207 0.00000908
## 4 208 0.00000908
## 5 209 0.00000908
## 6 210 0.00000908
## 7 211 0.00000908
## 8 212 0.00000908
## 9 213 0.00000908
## 10 214 0.00000908
## # ℹ 696 more rowsConversion between transmittance, absorptance and absorbance is also possible. In package ‘photobiology’ fractions of one are used to express transmittance, reflectance and absorptance.
any2Afr(filters.mspct$Schott_UG11, action = "replace")## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## Afr: Spectral absorptance [/1]
## --
## # A tibble: 706 × 2
## w.length Afr
## <dbl> <dbl>
## 1 200 1.000
## 2 204 1.000
## 3 207 1.000
## 4 208 1.000
## 5 209 1.000
## 6 210 1.000
## 7 211 1.000
## 8 212 1.000
## 9 213 1.000
## 10 214 1.000
## # ℹ 696 more rowsFor expressing absorbance, base-10 logarithms are used. In some
fields, natural logarithms are used instead. Expressing A
on this base is not supported by this package and any input absorbances
must be first converted to log10 based A.
any2A(filters.mspct$Schott_UG11, action = "replace")## Object: filter_spct [706 x 2]
## Wavelength range 200-5200 nm, step 1-50 nm
## Label: SCHOTT UG11 1mm-thick
## Rfr (/1): 0.092, thickness (mm): 1.00, attenuation mode: absorption.
## Variables:
## w.length: Wavelength [nm]
## A: Spectral absorbance log10 based [a.u.]
## --
## # A tibble: 706 × 2
## w.length A
## <dbl> <dbl>
## 1 200 5
## 2 204 5
## 3 207 5
## 4 208 5
## 5 209 5
## 6 210 5
## 7 211 5
## 8 212 5
## 9 213 5
## 10 214 5
## # ℹ 696 more rowsPlotting
Spectra can be plotted in the same ways as other data stored in data frames, using base R graphics, package ‘lattice’ or ‘ggplot2’. However, another package in our suite, ‘ggspectra’, built as an extension to ‘ggplot2’ makes plotting spectra extremely easy.
autoplot() methods use the metadata in the objects to
set labels and decorations, as well as automatically setting the mapping
of the x and y aesthetics.
autoplot(filters.mspct$MIDOPT_LP500)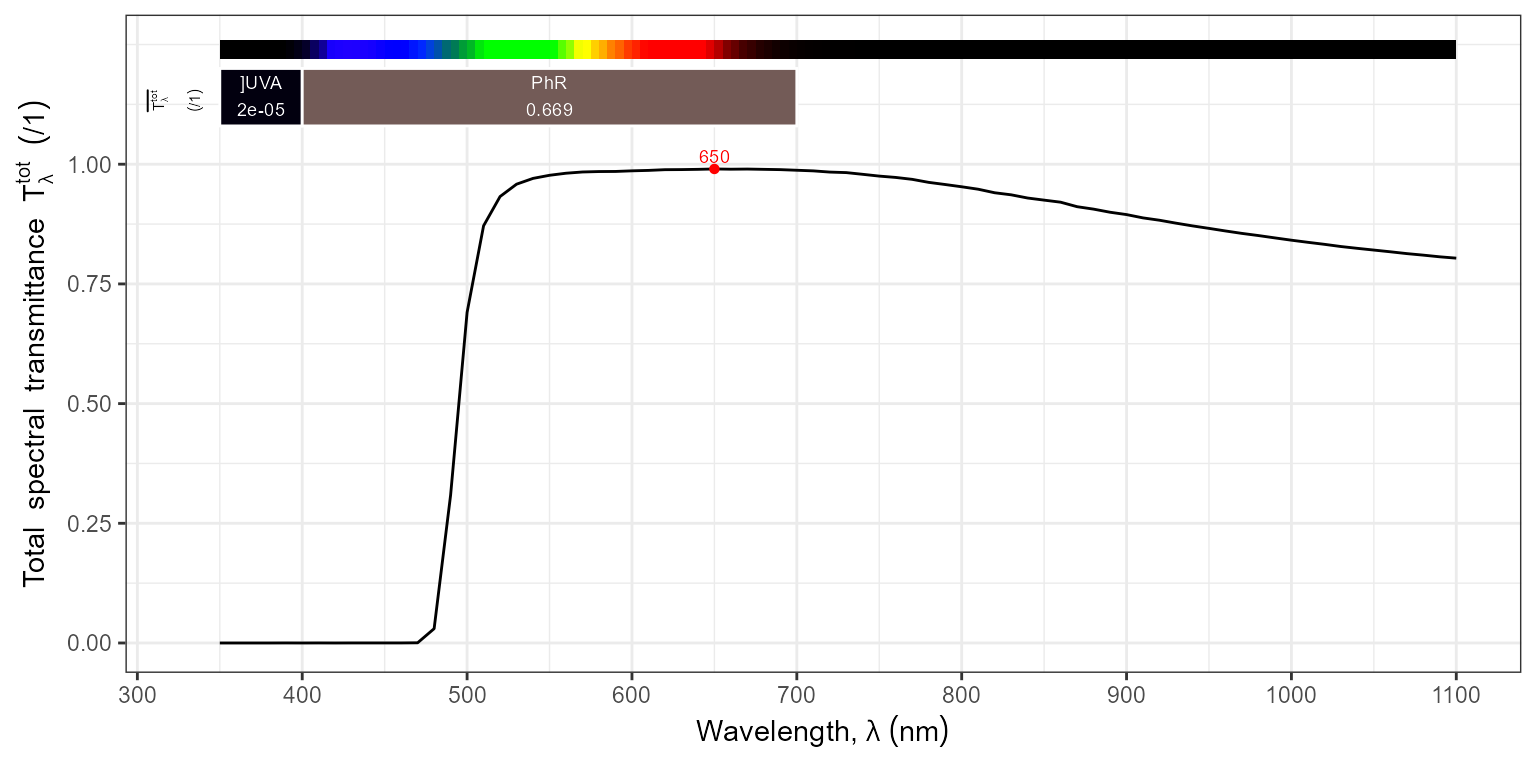
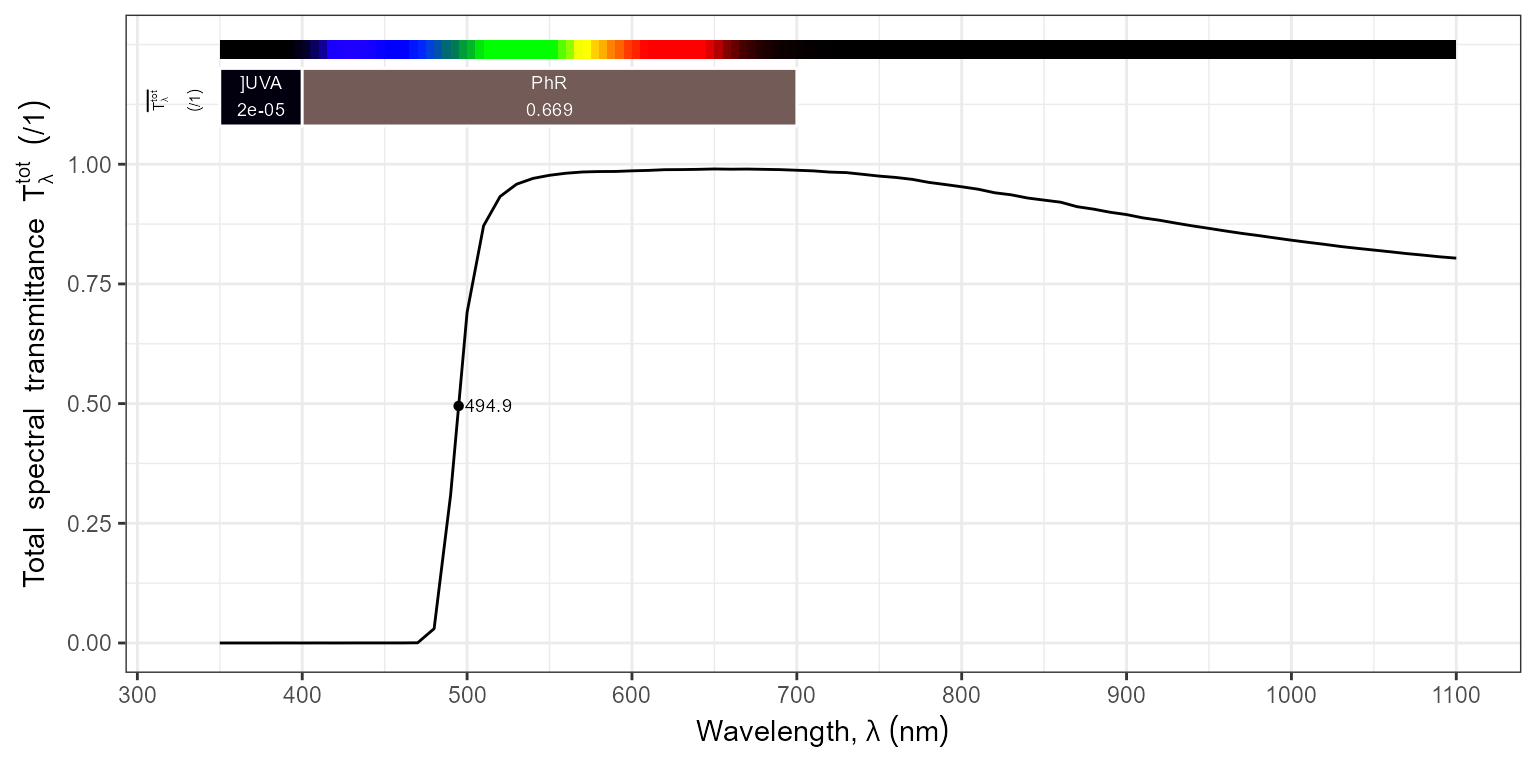
autoplot(filters.mspct$MIDOPT_TB550_660_850,
annotations = c("+", "title:none:none:what", "wls"),
w.band = VIS_bands(),
range = c(500, 910),
span = 11)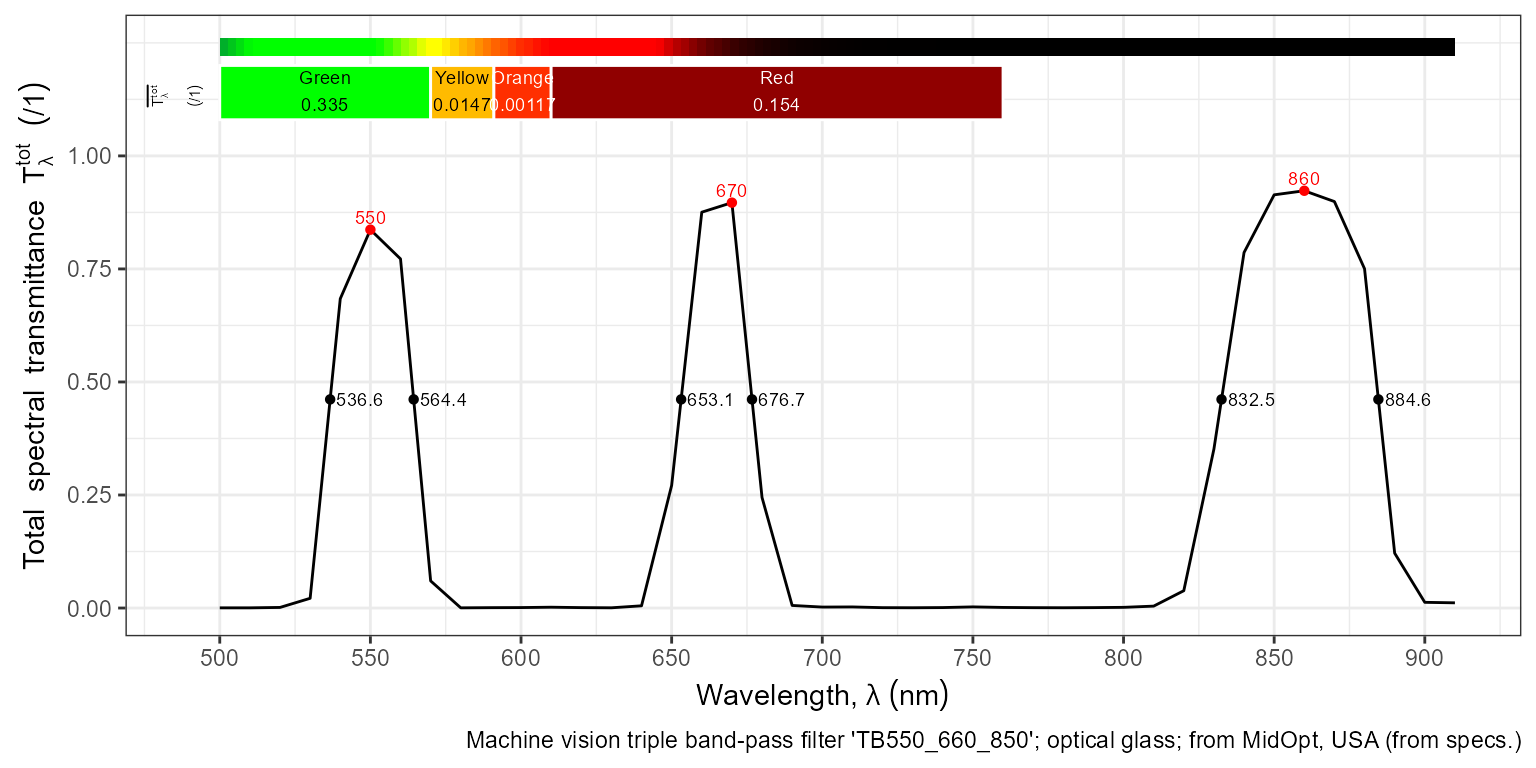
To graphically compare filters, we can pass a collection of spectral
objects, such as subset of filters.mspct.
autoplot(filters.mspct[c("Schott_UG1", "Schott_UG11")],
range = c(200, 900),
annotations = c("+", "boundaries"),
span = 11)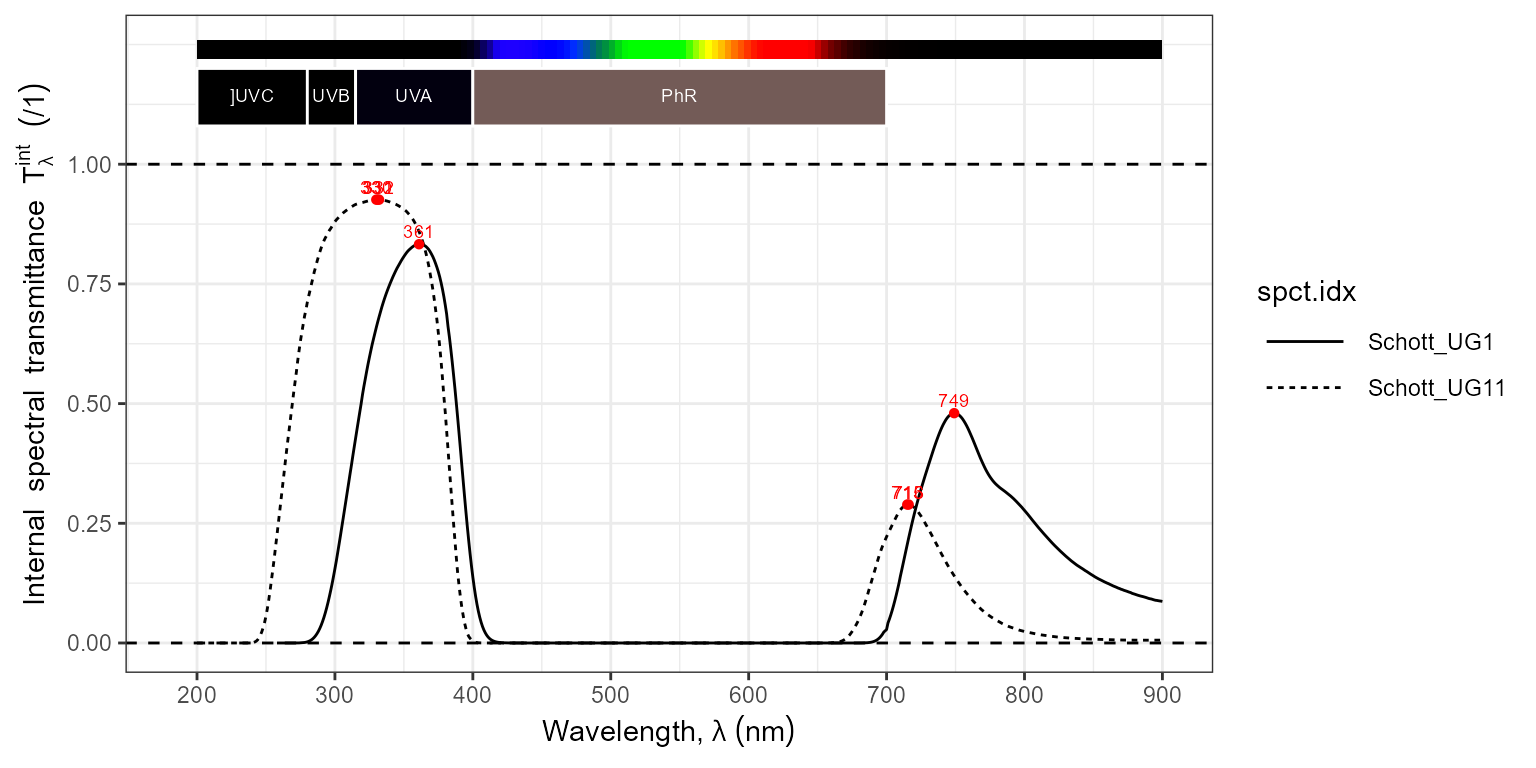
To graphichaly compare filter thicknesses we can pass a collection of spectral objects.
thin_and_thick.mspct <-
filter_mspct(list("1 mm" = filters.mspct$Schott_UG11,
"3 mm" = convertThickness(filters.mspct$Schott_UG11,
thickness = 3e-3)))
autoplot(thin_and_thick.mspct,
range = c(200, 900),
annotations = c("+", "boundaries"),
span = 101)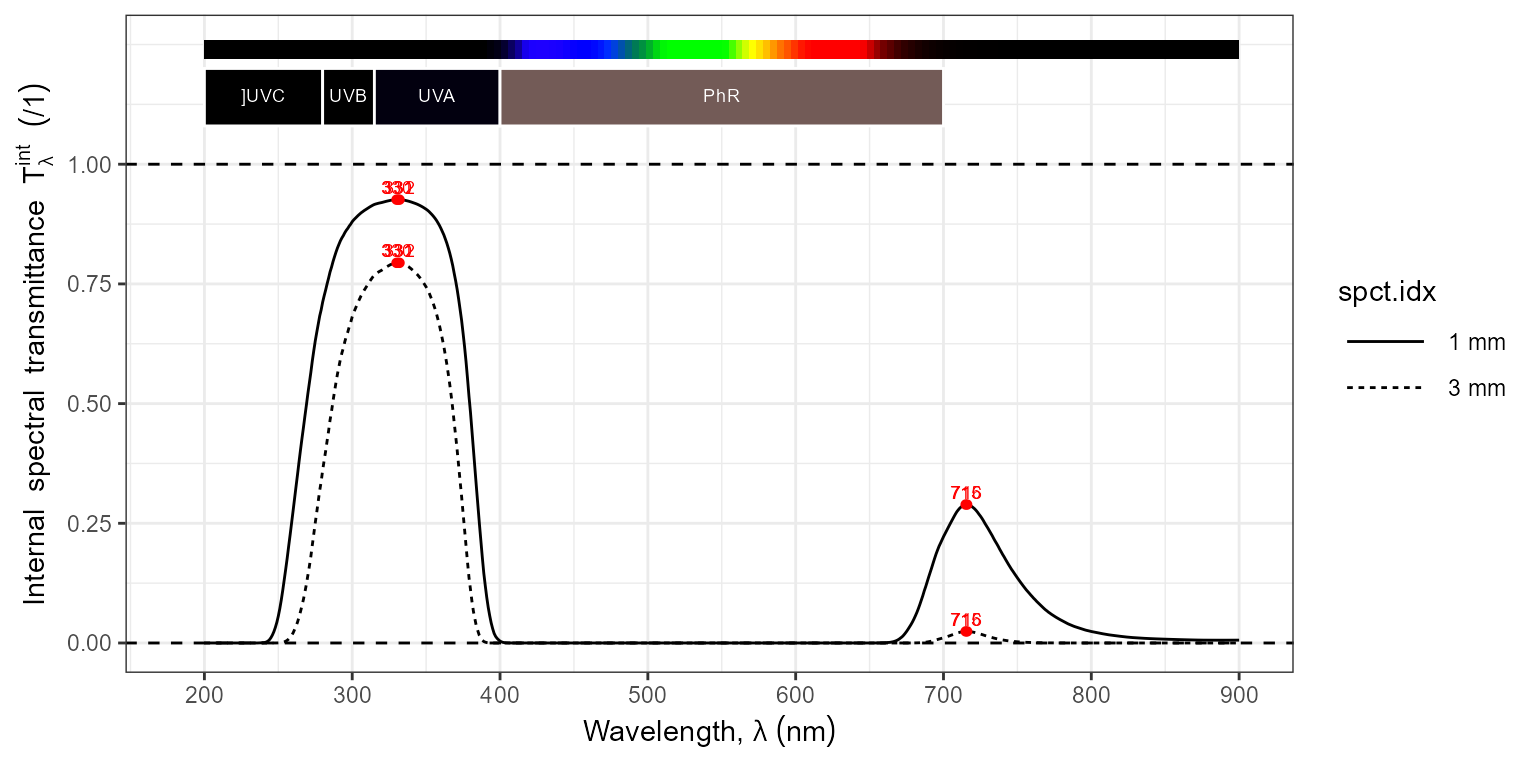
To graphically assess filter stacks with air gaps.
stack.spct <- filters.mspct$Haida_Clear_Night_NanoPro_1.6mm_52mm * filters.mspct$Firecrest_UVIR_Cut_0.96mm_52mm
autoplot(stack.spct,
range = c(NA, 1400),
w.band = c(UV_bands(), IR_bands("CIE")),
annotations = list(c("+", "boundaries"), c("-", "peaks")),
span = 21) +
geom_line(data = filters.mspct$Haida_Clear_Night_NanoPro_1.6mm_52mm, colour = "purple") +
geom_vline(xintercept = c(589, 589.6), linetype = "dotted") # Na emission lines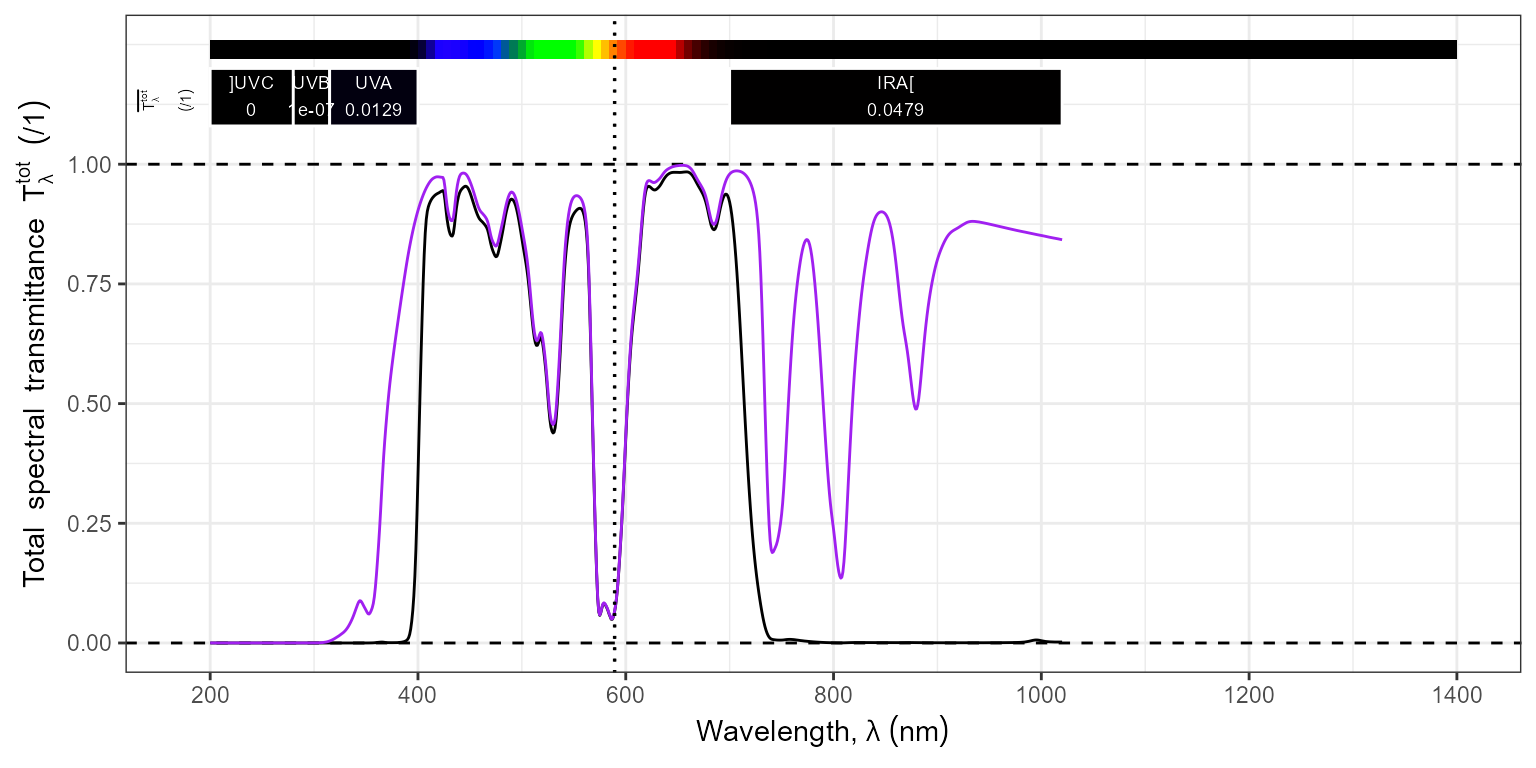
Package ‘ggspectra’ also defines specializations of method
ggplot() for spectra that automatically maps the
and
aesthetics.
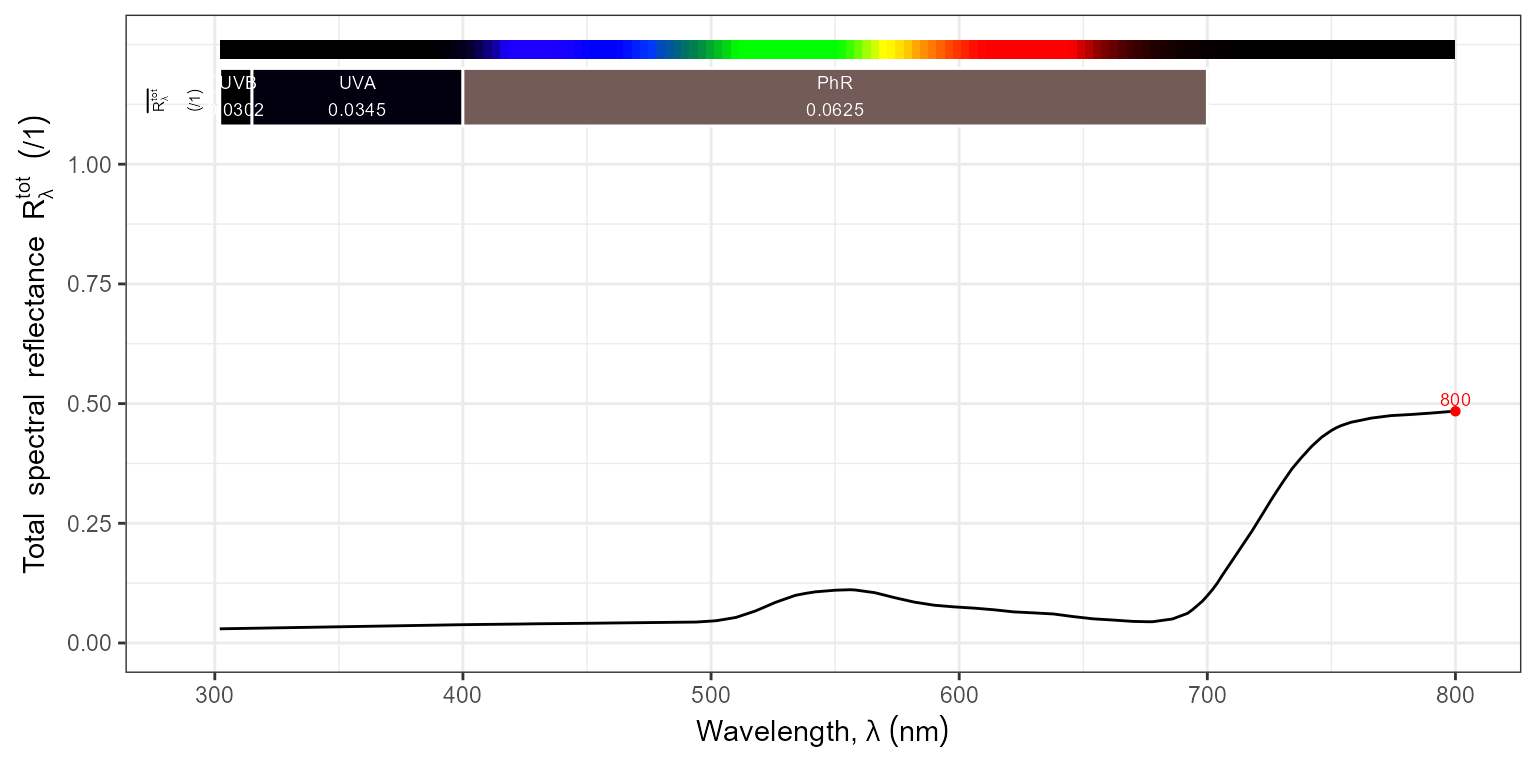
ggplot(filters.mspct$Firecrest_UVIR_Cut_0.96mm_52mm) +
geom_line() +
scale_x_wl_continuous() +
scale_y_Tfr_total_continuous()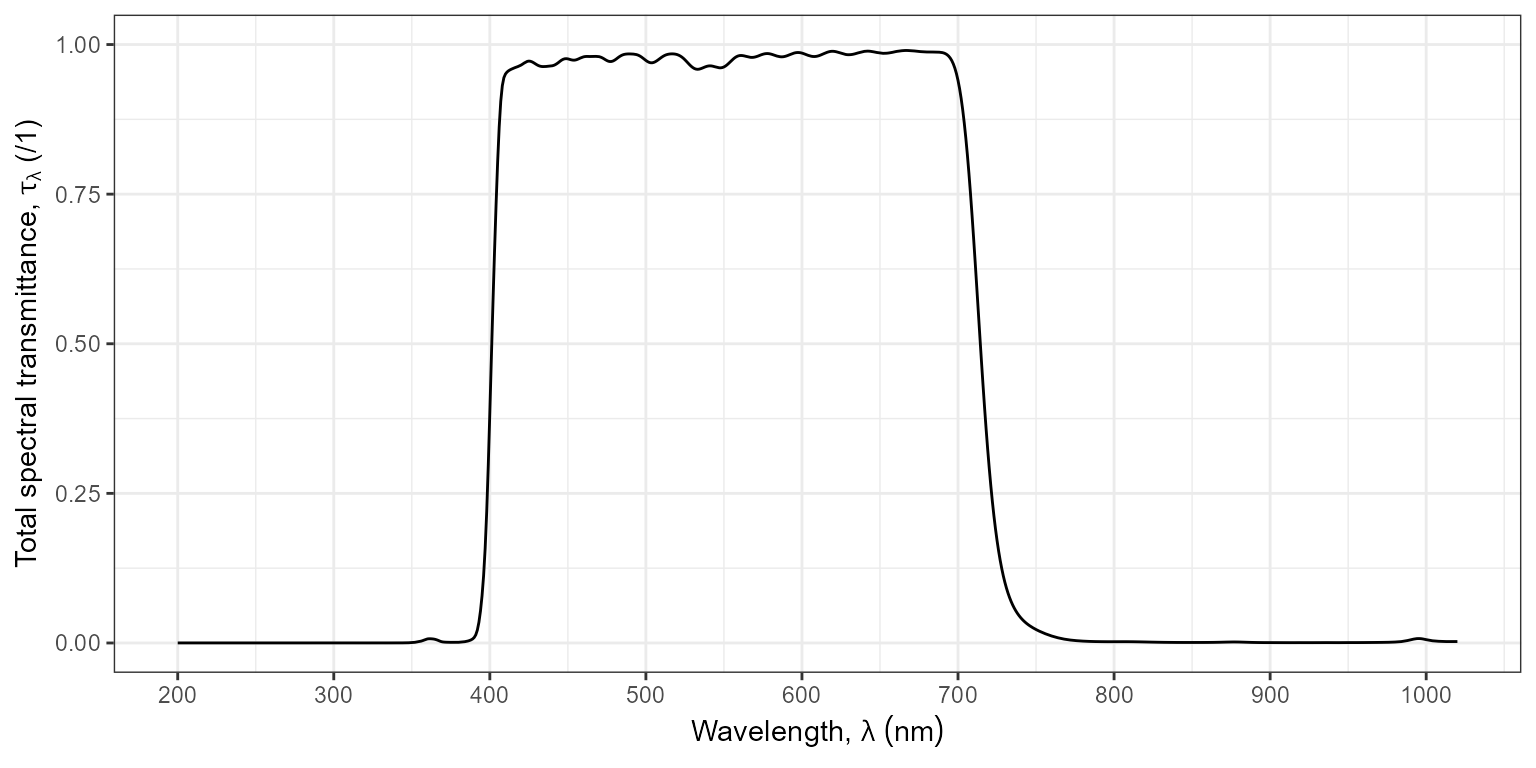
Calculating summaries
Here we use waveband defintions from package ‘photobiologyWavebands’ and summary functions from package ‘photobiology’.
transmittance(filters.mspct$Firecrest_UVIR_Cut_0.96mm_52mm, UVA())## Tfr(wl)_UVA.ISO
## 0.01554623
## attr(,"Tfr.type")
## [1] "total"
## attr(,"radiation.unit")
## [1] "transmittance average"
absorbance(filters.mspct$Firecrest_UVIR_Cut_0.96mm_52mm, list(UVA(), NIR()))## A(wl)_UVA.ISO A(wl)_NIR.ISO[
## 2.941898 2.898525
## attr(,"Tfr.type")
## [1] "total"
## attr(,"radiation.unit")
## [1] "absorbance average"## # A tibble: 5 × 3
## spct.idx `Tfr(wl)_UVB.ISO` `Tfr(wl)_UVA.ISO`
## <fct> <dbl> <dbl>
## 1 Schott_UG1 0.153 0.659
## 2 Schott_UG11 0.851 0.694
## 3 Schott_UG2A 0.0472 0.660
## 4 Schott_UG5 0.957 0.924
## 5 UQG_UG1_2mm 0.128 0.516
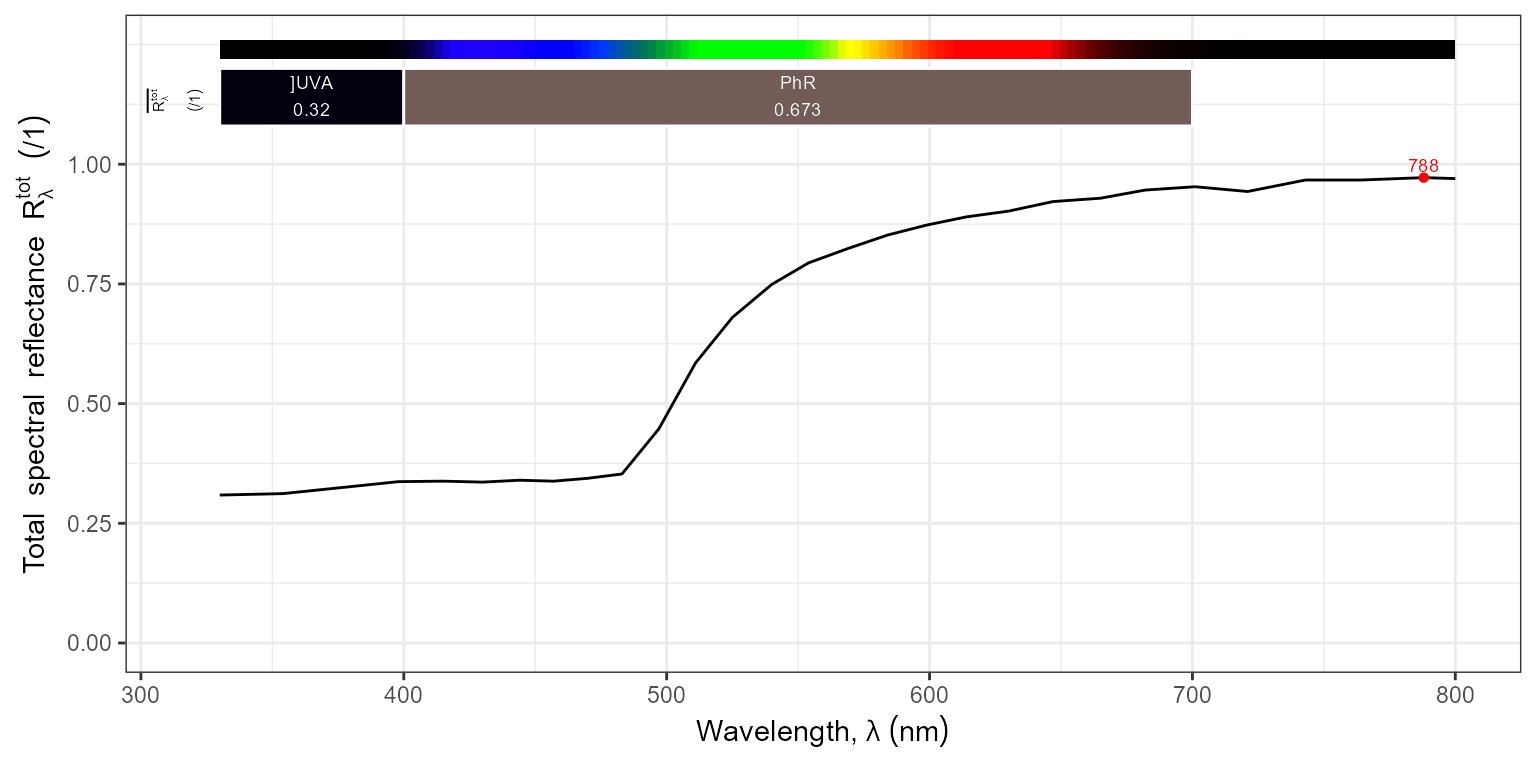
reflectance(metals.mspct, w.band = VIS_bands())## # A tibble: 12 × 7
## spct.idx `Rfr(wl)_Purple.ISO` `Rfr(wl)_Blue.ISO` `Rfr(wl)_Green.ISO`
## <fct> <dbl> <dbl> <dbl>
## 1 aluminum 0.843 0.865 0.866
## 2 copper 0.276 0.382 0.476
## 3 gold 0.333 0.366 0.702
## 4 molybdenum 0.444 0.500 0.508
## 5 nickel 0.445 0.523 0.563
## 6 platinum 0.612 0.666 0.715
## 7 silver 0.753 0.851 0.899
## 8 stainless_steel 0.526 0.579 0.603
## 9 tantalum 0.411 0.403 0.389
## 10 tin 0.701 0.797 0.822
## 11 titanium 0.166 0.202 0.222
## 12 vanadium 0.447 0.472 0.471
## # ℹ 3 more variables: `Rfr(wl)_Yellow.ISO` <dbl>, `Rfr(wl)_Orange.ISO` <dbl>,
## # `Rfr(wl)_Red.ISO` <dbl>Using the data in other contexts
As filter_mspct is a class derived from
list, and filter_spct is derived from
tibble::tible which is a mostly compatible reimplementation
of data.frame the data can be used very easily with any R
function.
head(as.data.frame(filters.mspct$Schott_UG11))## w.length Tfr
## 1 200 1e-05
## 2 204 1e-05
## 3 207 1e-05
## 4 208 1e-05
## 5 209 1e-05
## 6 210 1e-05Of course attach and with also work as
expected.
attach(filters.mspct)
transmittance(Schott_UG11, UVA())## Tfr(wl)_UVA.ISO
## 0.6942081
## attr(,"Tfr.type")
## [1] "internal"
## attr(,"radiation.unit")
## [1] "transmittance average"
detach(filters.mspct)## [1] 200 5200
detach(filters.mspct)
with(filters.mspct, transmittance(Schott_UG11, UVA()))## Tfr(wl)_UVA.ISO
## 0.6942081
## attr(,"Tfr.type")
## [1] "internal"
## attr(,"radiation.unit")
## [1] "transmittance average"