
Equation, p-value, \(R^2\), AIC and BIC of fitted polynomial
Source:R/stat-poly-eq.R
stat_poly_eq.Rdstat_poly_eq fits a polynomial, by default with stats::lm(),
but alternatively using robust, resistant or generalized least squares. Major
axis regression and segmented linear regression are also supported. Using the
fitted model it generates several labels including the fitted model equation,
p-value, F-value, coefficient of determination (R^2) and its confidence
interval, 'AIC', 'BIC', number of observations and method name, if available.
Usage
stat_poly_eq(
mapping = NULL,
data = NULL,
geom = "text_npc",
position = "identity",
...,
formula = NULL,
method = "lm",
method.args = list(),
n.min = 2L,
fit.seed = NA,
eq.with.lhs = TRUE,
eq.x.rhs = NULL,
small.r = getOption("ggpmisc.small.r", default = FALSE),
small.p = getOption("ggpmisc.small.p", default = FALSE),
CI.brackets = c("[", "]"),
rsquared.conf.level = 0.95,
coef.digits = 3,
coef.keep.zeros = TRUE,
decreasing = getOption("ggpmisc.decreasing.poly.eq", FALSE),
rr.digits = 2,
f.digits = 3,
p.digits = 3,
label.x = "left",
label.y = "top",
hstep = 0,
vstep = NULL,
output.type = NULL,
na.rm = FALSE,
orientation = NA,
parse = NULL,
show.legend = FALSE,
inherit.aes = TRUE
)Arguments
- mapping
The aesthetic mapping, usually constructed with
aes. Only needs to be set at the layer level if you are overriding the plot defaults.- data
A layer specific dataset, only needed if you want to override the plot defaults.
- geom
The geometric object to use display the data
- position
The position adjustment to use for overlapping points on this layer.
- ...
other arguments passed on to
layer. This can include aesthetics whose values you want to set, not map. Seelayerfor more details.- formula
a formula object. Using aesthetic names
xandyinstead of original variable names.- method
function or character If character, "lm", "rlm", "lqs". "gls" "ma", "sma", or the name of a model fit function are accepted, possibly followed by the fit function's
methodargument separated by a colon (e.g."rlm:M"). If a function is different tolm(),rlm(),lqs(),gls(),ma,sma, it must have formal parameters namedformula,data,weights, andmethod. See Details.- method.args
named list with additional arguments. Not
dataorweightswhich are always passed through aesthetic mappings.- n.min
integer Minimum number of distinct values in the explanatory variable (on the rhs of formula) for fitting to the attempted.
- fit.seed
RNG seed argument passed to
set.seed(). Defaults toNA, indicating thatset.seed()should not be called.- eq.with.lhs
If
characterthe string is pasted to the front of the equation label before parsing or alogical(see note).- eq.x.rhs
characterthis string will be used as replacement for"x"in the model equation when generating the label before parsing it.- small.r, small.p
logical Flags to switch use of lower case r and p for coefficient of determination and p-value.
- CI.brackets
character vector of length 2. The opening and closing brackets used for the CI label.
- rsquared.conf.level
numeric Confidence level for the returned confidence interval. Set to NA to skip CI computation.
- coef.digits, f.digits
integer Number of significant digits to use for the fitted coefficients and F-value.
- coef.keep.zeros
logical Keep or drop trailing zeros when formatting the fitted coefficients and F-value.
- decreasing
logical It specifies the order of the terms in the returned character string; in increasing (default) or decreasing powers.
- rr.digits, p.digits
integer Number of digits after the decimal point to use for \(R^2\) and P-value in labels. If
Inf, use exponential notation with three decimal places.- label.x, label.y
numericwith range 0..1 "normalized parent coordinates" (npc units) or character if usinggeom_text_npc()orgeom_label_npc(). If usinggeom_text()orgeom_label()numeric in native data units. If too short they will be recycled.- hstep, vstep
numeric in npc units, the horizontal and vertical step used between labels for different groups.
- output.type
character One of "expression", "LaTeX", "text", "markdown" or "numeric".
- na.rm
a logical indicating whether NA values should be stripped before the computation proceeds.
- orientation
character Either "x" or "y" controlling the default for
formula. The letter indicates the aesthetic considered the explanatory variable in the model fit.- parse
logical Passed to the geom. If
TRUE, the labels will be parsed into expressions and displayed as described in?plotmath. Default isTRUEifoutput.type = "expression"andFALSEotherwise.- show.legend
logical. Should this layer be included in the legends?
NA, the default, includes if any aesthetics are mapped.FALSEnever includes, andTRUEalways includes.- inherit.aes
If
FALSE, overrides the default aesthetics, rather than combining with them. This is most useful for helper functions that define both data and aesthetics and shouldn't inherit behaviour from the default plot specification, e.g.borders.
Value
A data frame, with a single row per group and columns as described
under Computed variables. In cases when the number of observations
is less than n.min a data frame with no rows or columns is returned,
and rendered as an empty/invisible plot layer.
Details
This statistic can be used to automatically annotate a plot with
\(R^2\), adjusted \(R^2\), the fitted model equation, \(P\), and
other parameters from a fitted model. It supports linear regression and
polynomial fits with lm(), segmented linear regression
with package 'segmented' and major axis and standardized major axis
regression with package 'smatr', robust and resistant regression with
packages 'MASS' and 'robustbase'. The list is not exhaustive, and depends
on the availability of methods for the model fit objects. Lack of methods
or explicit support results in individual parameters and matching labels
being set to NA. As some model fitting results can depend on the RNG,
fit.seed if different to NA is used as argument in a call to
set.seed() immediately ahead of model fitting.
While strings for \(R^2\), adjusted \(R^2\), \(F\), and \(P\)
annotations are returned for all valid linear models, A character string
for the fitted model is returned only for polynomials (see below). When
not generated automatically, the equation can still be assembled by the
user within the call to aes(). In addition, a label
for the confidence interval of \(R^2\), based on values computed with
function ci_rsquared from package 'confintr' is
returned when possible.
Model formulas can use poly() or be defined algebraically including
the intercept indicated by +1, -1, +0 or implicit. If
defined using poly() the argument raw = TRUE must be passed.
The model formula is checked, and if not recognized as a polynomial
with no missing terms and terms ordered by increasing powers, no equation
label is generated. Thus, as the value returned for eq.label can be
NA, the default aesthetic mapping to label is \(R^2\).
The character strings mapped to the label aesthetic are encoded
differently depending on argument passed to output.type, or
if none passed based on the geom used. The argument of
parse is set automatically based on output.type. However,
if labels manually assembled from numeric output need parsing,
the default needs to be overridden.
This statistic only generates annotation labels, the predicted values/line
need to be added to the plot as a separate layer using
stat_poly_line (or stat_smooth).
Passing the same arguments in stat_poly_line() and in
stat_poly_eq() to parameters method and formula, and
if used also to method.args ensures that the plotted curve and
equation are consistent. Thus, it is best to save these arguments as named
objects and pass them as arguments to the two statistics.
A ggplot statistic receives as data a data frame that is not the one
passed as argument by the user, but instead a data frame with the variables
mapped to aesthetics. stat_poly_eq() mimics how
stat_smooth() works. Thus, the model formula should
be defined based on the names of aesthetics x and y, not the
names of the variables in the data. Before fitting the model, data are
split based on groupings created by any other
mappings present in a plot panel: fitting is done separately for each
group in each plot panel.
With method "lm", singularity results in terms being dropped with a
message if more numerous than can be fitted with a singular (exact) fit. In
this case or if the model results in a perfect fit due to a low number of
observations, estimates for various parameters are NaN or NA.
When this is the case the corresponding labels are set to
character(0L) and thus not visible in the plot. With methods other
than "lm", the model fit functions simply fail in case of
singularity, e.g., singular fits are not implemented in
rlm().
A requirement for a minimum number of observations with distinct values in
the explanatory variable can be set through parameter n.min. The
default n.min = 2L is the smallest suitable for method "lm"
but too small for method "rlm" for which n.min = 3L is
needed. Anyway, model fits with very few observations are of little
interest and using larger values of n.min than the default is
usually wise. This can be useful as when this threshold is not reached
an empty data frame is returned resulting in an empty plot layer.
R option OutDec is obeyed based on its value at the time the plot
is rendered, i.e., displayed or printed. Set options(OutDec = ",")
for languages like Spanish or French.
Note
stat_poly_eq() understands x and y,
to be referenced in the formula and weight passed as argument
to parameter weights. All three must be mapped to numeric
variables.
If the model formula includes a transformation of x, a
matching argument should be passed to parameter eq.x.rhs
as its default value "x" will not reflect the applied
transformation. In plots, transformation should never be applied to the
left hand side of the model formula, but instead in the mapping of the
variable within aes, as otherwise plotted observations and fitted
curve will not match. In this case it may be necessary to also pass
a matching argument to parameter eq.with.lhs.
For backward compatibility a logical is accepted as argument for
eq.with.lhs. If TRUE, the default is used, either
"x" or "y", depending on the argument passed to formula.
However, "x" or "y" can be substituted by providing a
suitable replacement character string through eq.x.rhs.
Parameter orientation is redundant as it only affects the default
for formula but is included for consistency with
ggplot2::stat_smooth().
User-defined methods
User-defined functions can be passed as
argument to method. The requirements are 1) that the signature is
similar to that of function lm() (with parameters formula,
data, weights and any other arguments passed by name through
method.args) and 2) that the value returned by the function is an
object of a class such as "lm" for which coefs() and similar
query methods are available or for empty plot layer output, an atomic
NA value.
When possible, i.e., nearly allways, the formula used to build
the equation label is extracted from the returned fitted model object.
Most fitted model objects returned follow the example of lm() and
include the model formula fitted. Thus, this model formula can safely
differ from the argument passed to parameter formula in the call
to stat_poly_eq().
Thus, user-defined methods can implement any or all of method
selection, model formula selection, dynamically adjusted
method.args and conditional skipping of labelling on a by group
basis.
Computed variables
If the model fit function used does not returns NA or no value,
the label is set to character(0L). The position of the columns in
the data frame can change between package versions, extract values always
by name.
For all output.type arguments the following values are returned.
- x,npcx
x position
- y,npcy
y position
- coefs
fitted coefficients, named numeric vector as a list member
- r.squared, rr.confint.level, rr.confint.low, rr.confint.high, adj.r.squared, f.value, f.df1, f.df2, p.value, AIC, BIC, n, knots, knots.se
numeric values, from the model fit object
- grp.label
Set according to mapping in
aes.- knots
list containing a numeric vector of knot or "psi" x-value for linear splines
- fm.method
name of method used, character
- fm.class
most derived class or the fitted model object, character
- fm.formula.chr
formatted model formula, character
If output.type is not "numeric" the returned tibble contains in
addition to those above the columns listed below, each containing a single
character string. The markup used depends on the value of output.type.
- eq.label
equation for the fitted polynomial as a character string to be parsed or
NA- rr.label
\(R^2\) of the fitted model as a character string to be parsed
- adj.rr.label
Adjusted \(R^2\) of the fitted model as a character string to be parsed
- rr.confint.label
Confidence interval for \(R^2\) of the fitted model as a character string to be parsed
- f.value.label
F value and degrees of freedom for the fitted model as a whole.
- p.value.label
P-value for the F-value above.
- AIC.label
AIC for the fitted model.
- BIC.label
BIC for the fitted model.
- n.label
Number of observations used in the fit.
- knots.label
The knots or change points in segmented regression.
- grp.label
Set according to mapping in
aes.- method.label
Set according
methodused.
If output.type is "numeric" the returned tibble contains columns
listed below in addition to the base ones. If the model fit function used
does not return a value, the variable is set to NA_real_.
- coef.ls
list containing the "coefficients" matrix from the summary of the fit object
- b_0.constant
TRUE is polynomial is forced through the origin
- b_i
One or more columns with the coefficient estimates
To explore the computed values returned for a given input we suggest the use
of geom_debug as shown in the last examples below.
Output types
The formatting of character strings to be displayed in plots are marked as mathematical equations. Depending on the geom used, the mark-up needs to be encoded differently, or in some cases mark-up not applied.
"expression"The labels are encoded as character strings to be parsed into R's plotmath expressions.
"LaTeX", "TeX", "tikz", "latex"The labels are encoded as 'LaTeX' maths equations, without the "fences" for switching in math mode.
"latex.eqn"Same as
"latex"but enclosed in single$, i.e., as in-line maths."latex.deqn"Same as
"latex"but enclosed in double$$, i.e., as display maths."markdown"The labels are encoded as character strings using markdown syntax, with some embedded HTML.
"marquee"The labels are encoded as character strings using markdown syntax, with 'marquee' supported spans.
"text"The labels are plain ASCII character strings.
"numeric"No labels are generated. This value is accepted by the statistics, but not by the label formatting functions.
NULLThe value used,
expression,latex.eqnormarkupdepends on the argument passed togeom.
If geom = "latex" (package 'xdvir') the output type used is
"latex.eqn". If geom = "richtext" (package 'ggtext') or
geom = "textbox" (package 'ggtext') the output type used is
"markdown". If geom = "marquee" (package 'marquee') the output
type used is "marquee". For all other values of geom the default
is "expression" unless the user passes an argument. Invalid values as
argument trigger an Error.
References
Originally written as an answer to question 7549694 at Stackoverflow but enhanced based on suggestions from users and my own needs.
See also
This statistics fits a model with function lm()
as default, several other functions returning objects of class "lm"
or objects of classes for which the common R fitted-model-object
extraction/query methods are available. Consult the documentation of these
functions for the details and additional arguments that can be passed to
them by name through parameter method.args. User-defined
model-fitting functions are also supported.
Please, see the articles online documentation for additional use examples and guidance.
For quantile regression stat_quant_eq() can be used
instead of stat_poly_eq() while for model II or major axis
regression with package 'lmodel2' stat_ma_eq() should be
used. For methods not supported by these three statistics, such as
non-linear models, statistics stat_fit_glance() and
stat_fit_tidy() can be used but require the user to create
character strings from numeric values and map them to aesthetic
label.
Other ggplot statistics for linear and polynomial regression:
stat_poly_line()
Aesthetics
stat_poly_eq() understands the following aesthetics. Required aesthetics are displayed in bold and defaults are displayed for optional aesthetics:
| • | x | |
| • | y | |
| • | group | → inferred |
| • | grp.label | |
| • | hjust | → "inward" |
| • | label | → after_stat(rr.label) |
| • | npcx | → after_stat(npcx) |
| • | npcy | → after_stat(npcy) |
| • | vjust | → "inward" |
| • | weight | → 1 |
Learn more about setting these aesthetics in vignette("ggplot2-specs").
Examples
# generate artificial data
set.seed(4321)
x <- 1:100
y <- (x + x^2 + x^3) + rnorm(length(x), mean = 0, sd = mean(x^3) / 4)
y <- y / max(y)
my.data <- data.frame(x = x, y = y,
group = c("A", "B"),
y2 = y * c(1, 2) + c(0, 0.1),
w = sqrt(x))
# give a name to a formula
formula <- y ~ poly(x, 3, raw = TRUE)
# using defaults
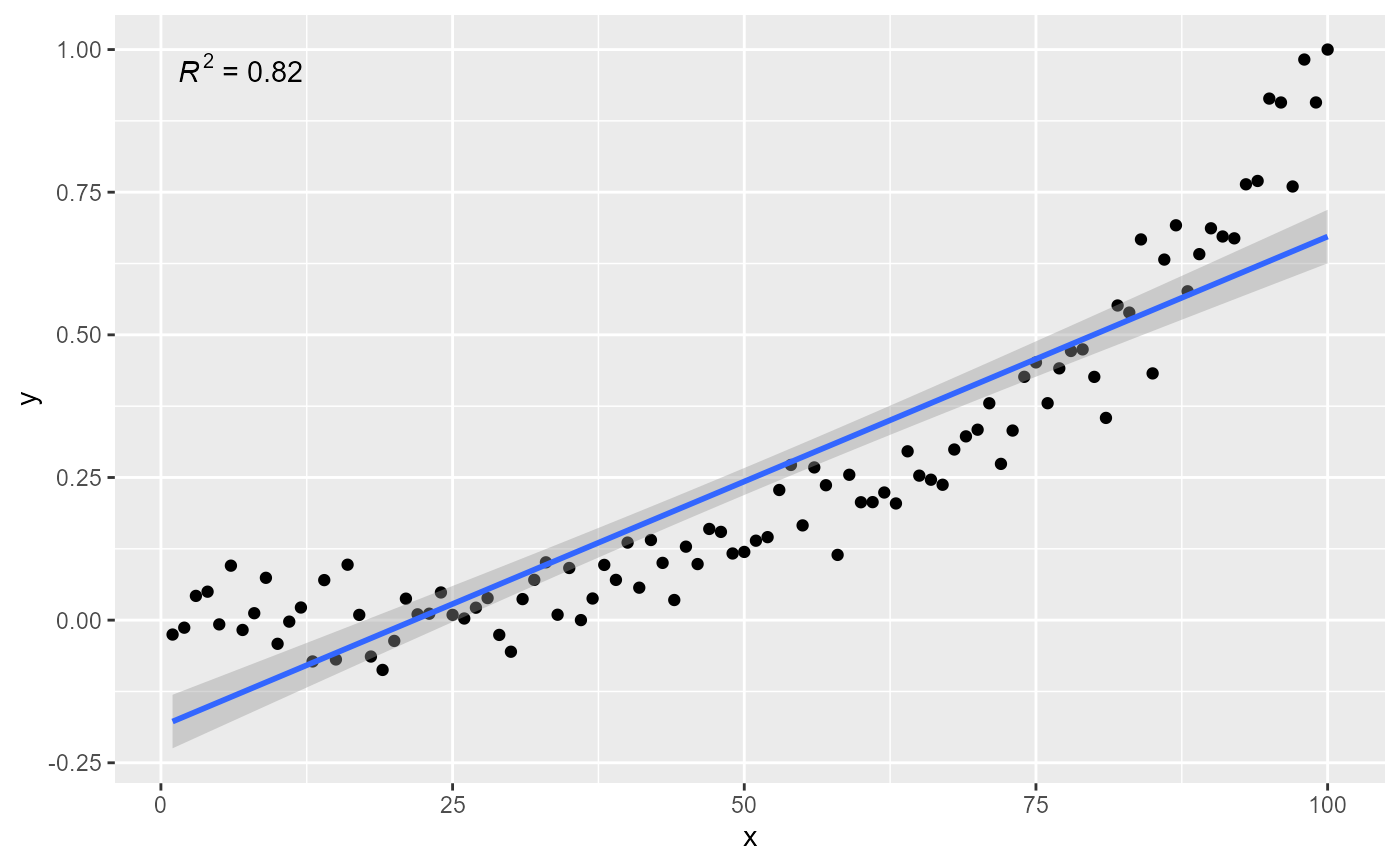
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line() +
stat_poly_eq()
 # no weights
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula)
# no weights
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula)
 # other labels
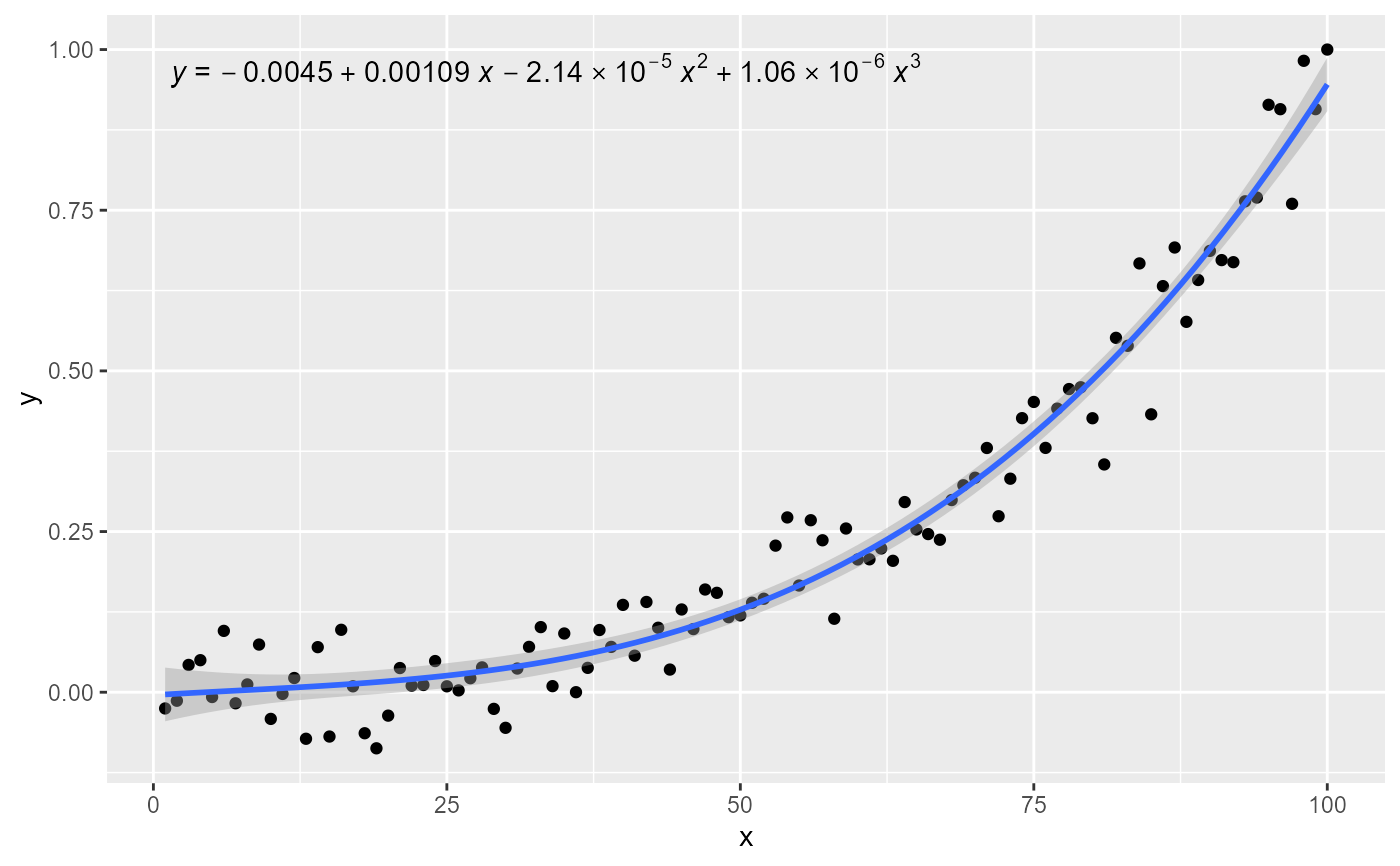
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("eq"), formula = formula)
# other labels
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("eq"), formula = formula)
 # other labels
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("eq"), formula = formula, decreasing = TRUE)
# other labels
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("eq"), formula = formula, decreasing = TRUE)
 ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("eq", "R2"), formula = formula)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("eq", "R2"), formula = formula)
 ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("R2", "R2.CI", "P", "method"), formula = formula)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("R2", "R2.CI", "P", "method"), formula = formula)
 ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("R2", "F", "P", "n", sep = "*\"; \"*"),
formula = formula)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(use_label("R2", "F", "P", "n", sep = "*\"; \"*"),
formula = formula)
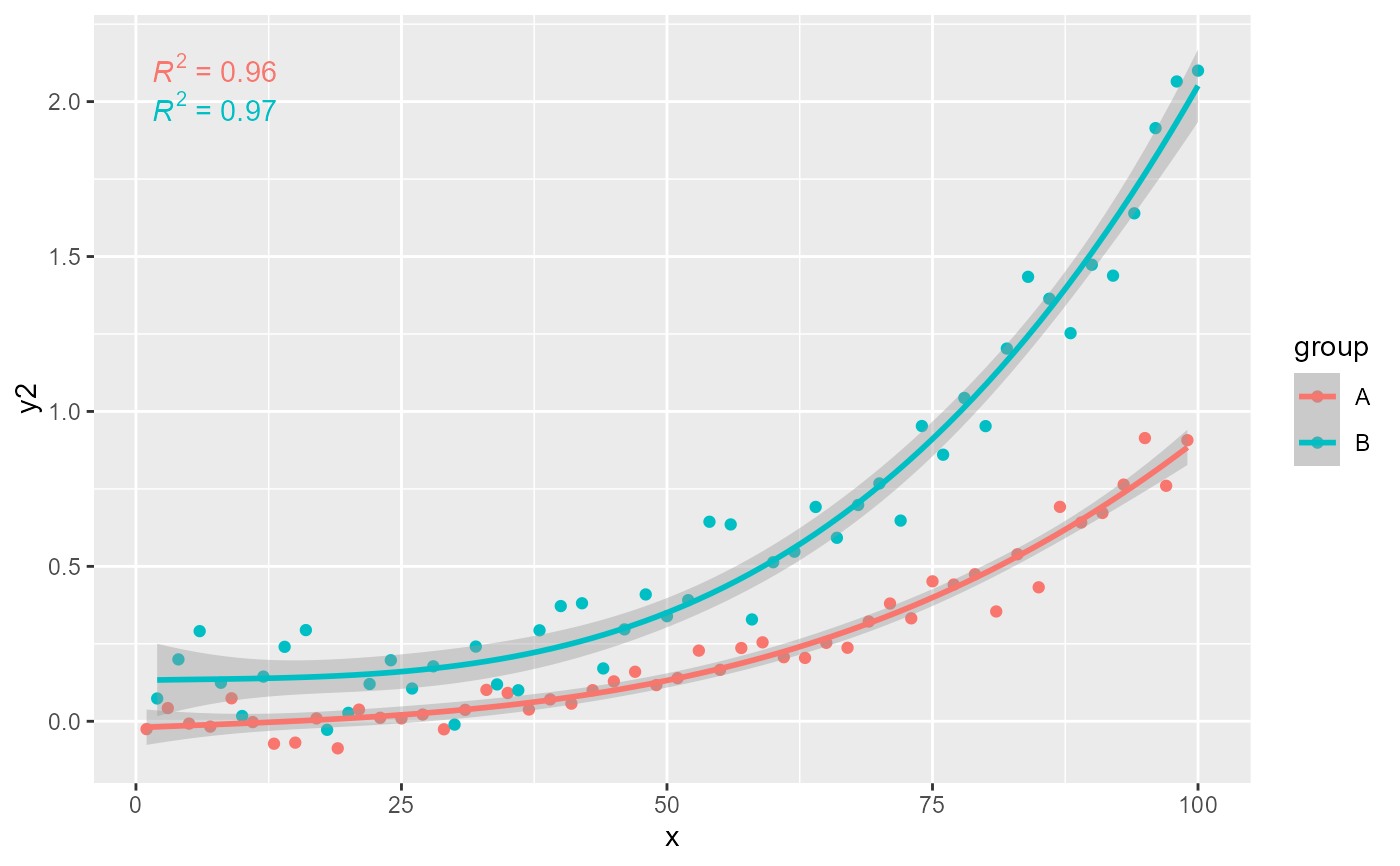 # grouping
ggplot(my.data, aes(x, y2, color = group)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula)
# grouping
ggplot(my.data, aes(x, y2, color = group)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula)
 # rotation
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, angle = 90)
# rotation
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, angle = 90)
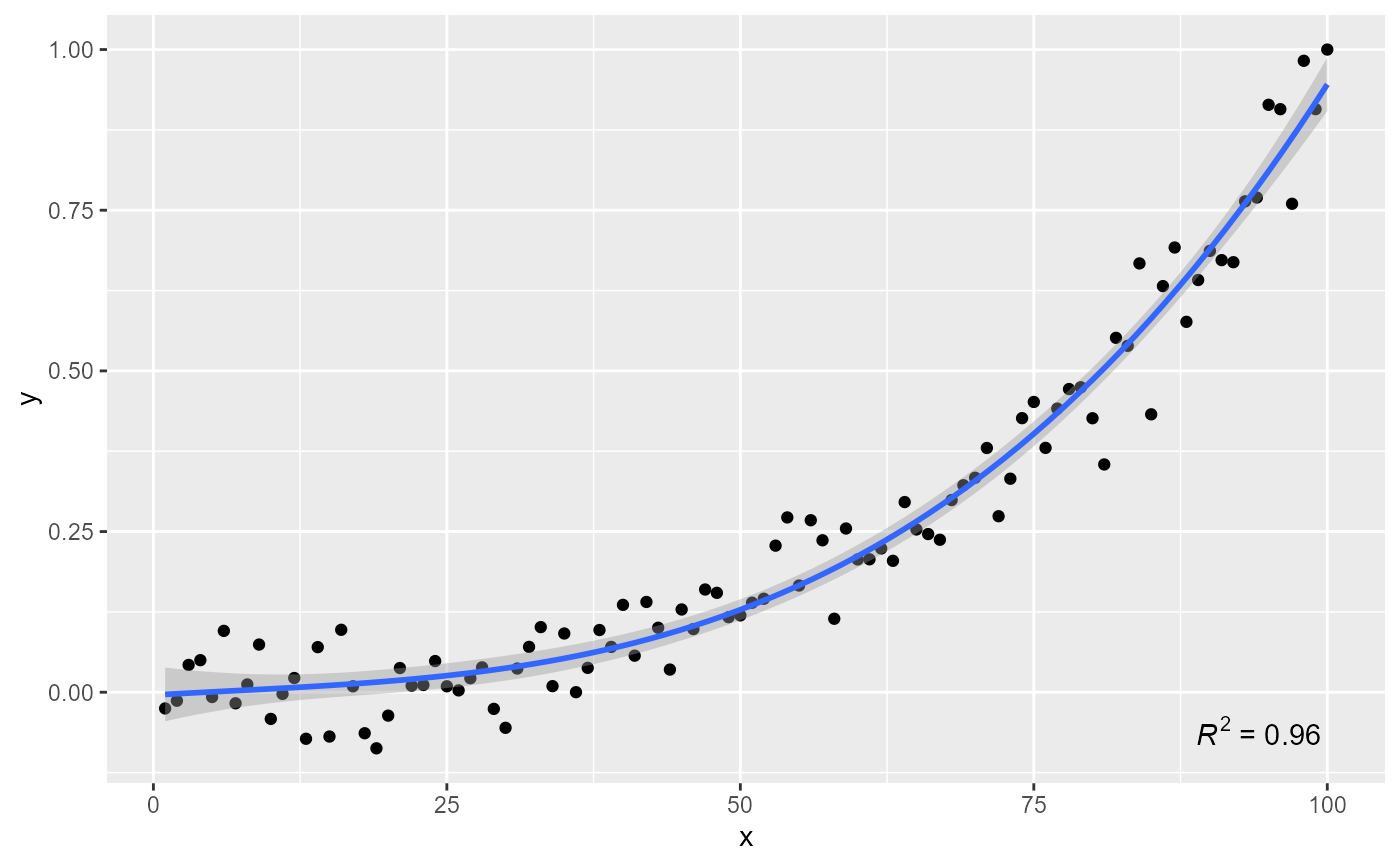 # label location
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, label.y = "bottom", label.x = "right")
# label location
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, label.y = "bottom", label.x = "right")
 ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, label.y = 0.1, label.x = 0.9)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, label.y = 0.1, label.x = 0.9)
 # modifying the explanatory variable within the model formula
# modifying the response variable within aes()
# eq.x.rhs and eq.with.lhs defaults must be overridden!!
formula.trans <- y ~ I(x^2)
ggplot(my.data, aes(x, y + 1)) +
geom_point() +
stat_poly_line(formula = formula.trans) +
stat_poly_eq(use_label("eq"),
formula = formula.trans,
eq.x.rhs = "~x^2",
eq.with.lhs = "y + 1~~`=`~~")
# modifying the explanatory variable within the model formula
# modifying the response variable within aes()
# eq.x.rhs and eq.with.lhs defaults must be overridden!!
formula.trans <- y ~ I(x^2)
ggplot(my.data, aes(x, y + 1)) +
geom_point() +
stat_poly_line(formula = formula.trans) +
stat_poly_eq(use_label("eq"),
formula = formula.trans,
eq.x.rhs = "~x^2",
eq.with.lhs = "y + 1~~`=`~~")
 # using weights
ggplot(my.data, aes(x, y, weight = w)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula)
# using weights
ggplot(my.data, aes(x, y, weight = w)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula)
 # no weights, 4 digits for R square
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, rr.digits = 4)
# no weights, 4 digits for R square
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula, rr.digits = 4)
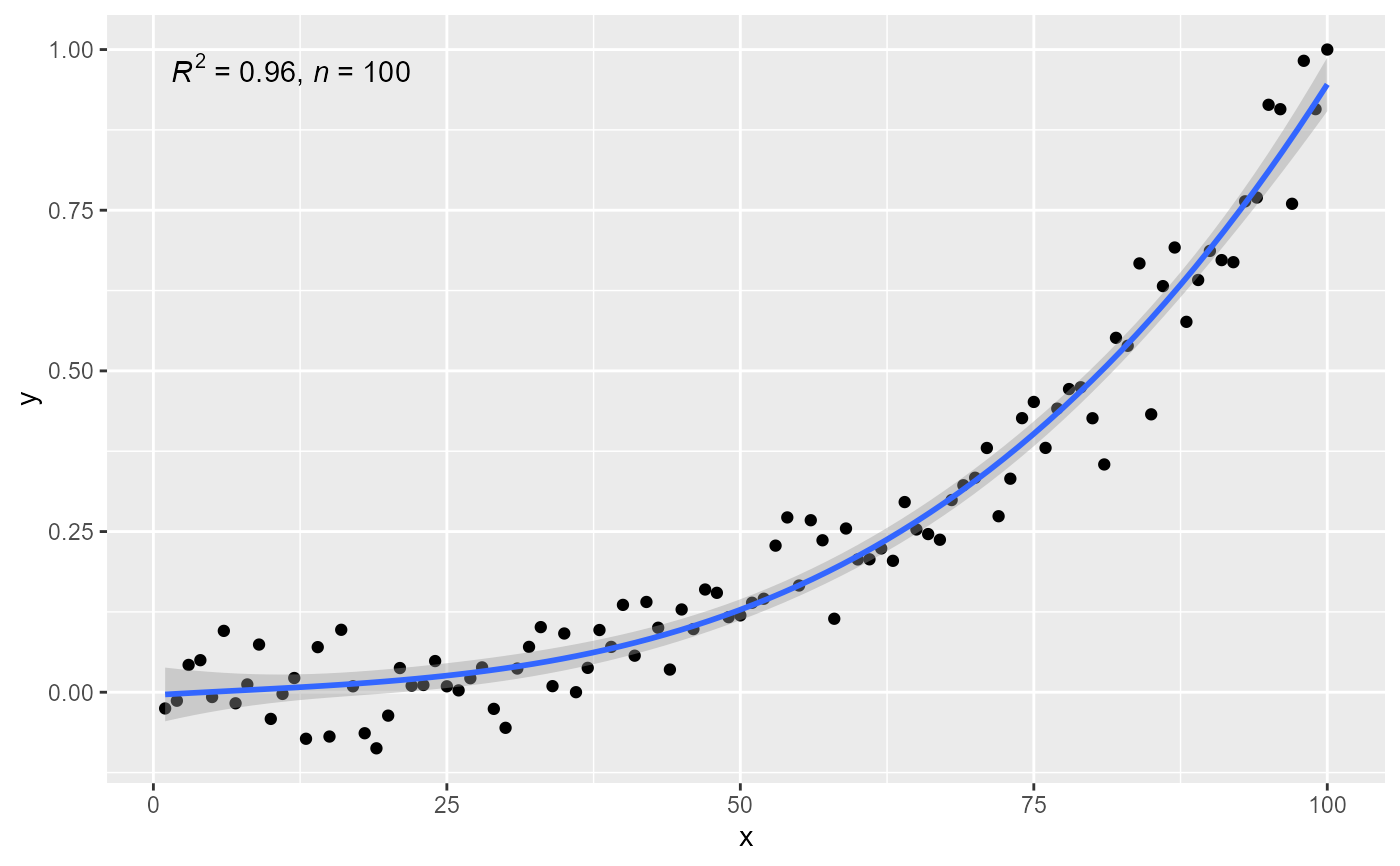 # manually assemble and map a specific label using paste() and aes()
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(aes(label = paste(after_stat(rr.label),
after_stat(n.label), sep = "*\", \"*")),
formula = formula)
# manually assemble and map a specific label using paste() and aes()
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(aes(label = paste(after_stat(rr.label),
after_stat(n.label), sep = "*\", \"*")),
formula = formula)
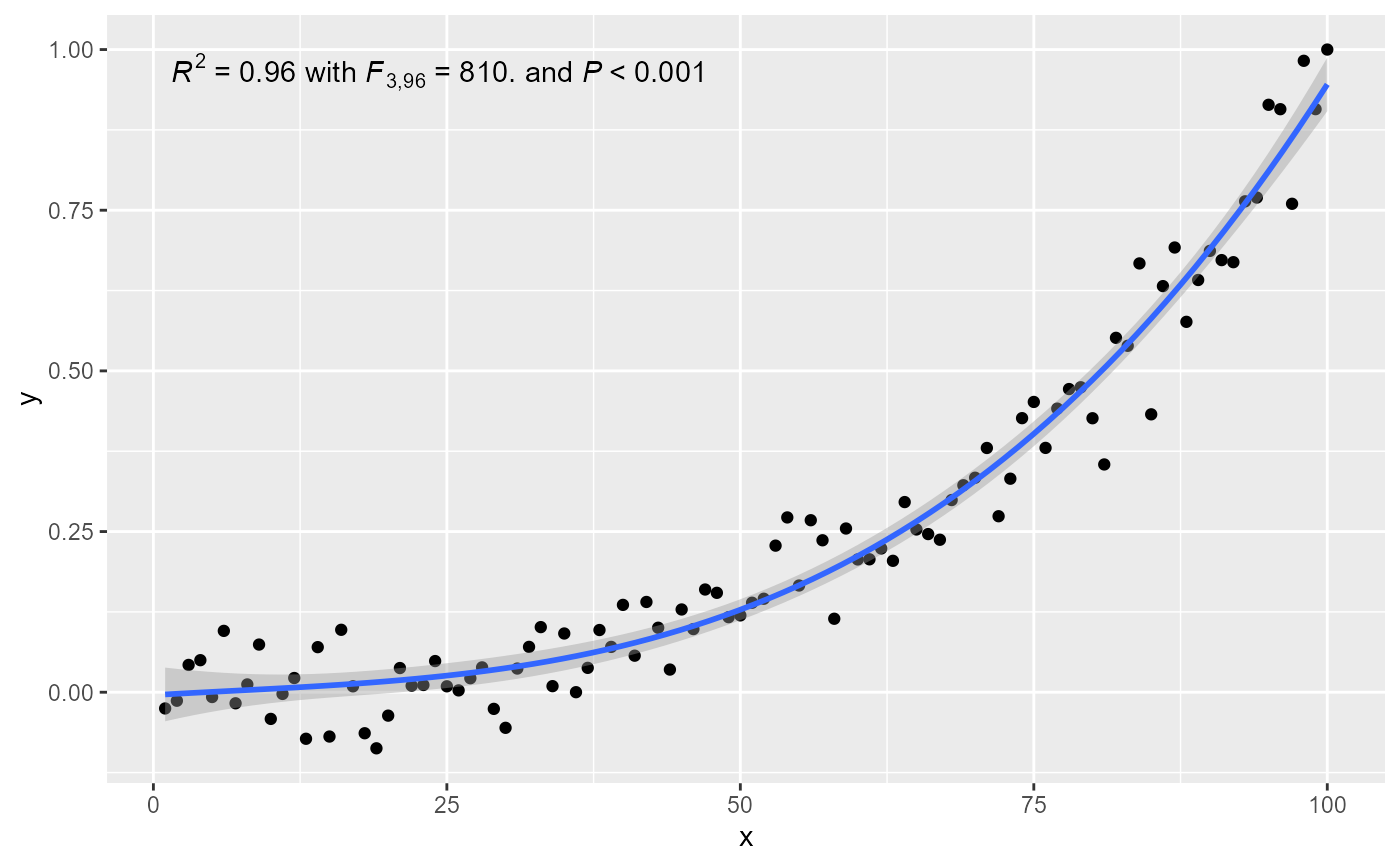 # manually assemble and map a specific label using sprintf() and aes()
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(aes(label = sprintf("%s*\" with \"*%s*\" and \"*%s",
after_stat(rr.label),
after_stat(f.value.label),
after_stat(p.value.label))),
formula = formula)
# manually assemble and map a specific label using sprintf() and aes()
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(aes(label = sprintf("%s*\" with \"*%s*\" and \"*%s",
after_stat(rr.label),
after_stat(f.value.label),
after_stat(p.value.label))),
formula = formula)
 # x on y regression
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula, orientation = "y") +
stat_poly_eq(use_label("eq", "adj.R2"),
formula = x ~ poly(y, 3, raw = TRUE))
# x on y regression
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula, orientation = "y") +
stat_poly_eq(use_label("eq", "adj.R2"),
formula = x ~ poly(y, 3, raw = TRUE))
 # conditional user specified label
ggplot(my.data, aes(x, y2, color = group)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(aes(label = ifelse(after_stat(adj.r.squared) > 0.96,
paste(after_stat(adj.rr.label),
after_stat(eq.label),
sep = "*\", \"*"),
after_stat(adj.rr.label))),
rr.digits = 3,
formula = formula)
# conditional user specified label
ggplot(my.data, aes(x, y2, color = group)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(aes(label = ifelse(after_stat(adj.r.squared) > 0.96,
paste(after_stat(adj.rr.label),
after_stat(eq.label),
sep = "*\", \"*"),
after_stat(adj.rr.label))),
rr.digits = 3,
formula = formula)
 # geom = "text"
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(geom = "text", label.x = 100, label.y = 0, hjust = 1,
formula = formula)
# geom = "text"
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(geom = "text", label.x = 100, label.y = 0, hjust = 1,
formula = formula)
 # Inspecting the returned data using geom_debug_group()
# This provides a quick way of finding out the names of the variables that
# are available for mapping to aesthetics with after_stat().
gginnards.installed <- requireNamespace("gginnards", quietly = TRUE)
if (gginnards.installed)
library(gginnards)
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group")
# Inspecting the returned data using geom_debug_group()
# This provides a quick way of finding out the names of the variables that
# are available for mapping to aesthetics with after_stat().
gginnards.installed <- requireNamespace("gginnards", quietly = TRUE)
if (gginnards.installed)
library(gginnards)
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group")
 #> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (head):"
#> eq.label
#> 1 italic(y)~`=`~-0.00450 + 0.00109*~italic(x) - 2.14%*% 10^{-05}*~italic(x)^2 + 1.06%*% 10^{-06}*~italic(x)^3
#> rr.label adj.rr.label rr.confint.label
#> 1 italic(R)^2~`=`~"0.96" italic(R)[adj]^2~`=`~"0.96" "95% CI [0.95, 0.97]"
#> AIC.label BIC.label
#> 1 plain(AIC)~`=`~"-291.9" plain(BIC)~`=`~"-278.8"
#> f.value.label p.value.label n.label
#> 1 italic(F)[3*","*96]~`=`~"810." italic(P)~`<`~"0.001" italic(n)~`=`~100
#> knots.label grp.label method.label
#> 1 <NA> -1 "method: lm:qr"
#> coefs
#> 1 -4.495631e-03, 1.089644e-03, -2.139937e-05, 1.055387e-06
#> coefs.names
#> 1 (Intercept), poly(x, 3, raw = TRUE)1, poly(x, 3, raw = TRUE)2, poly(x, 3, raw = TRUE)3
#> rr.confint.level rr.confint.low rr.confint.high f.value f.df1 f.df2 r.squared
#> 1 0.95 0.9464423 0.9696371 810.484 3 96 0.9620171
#> adj.r.squared p.value AIC BIC n knots knots.se knots.names
#> 1 0.9608301 5.110349e-68 -291.8639 -278.838 100 NA NA NA
#> fm.method fm.class fm.formula fm.formula.chr x
#> 1 lm:qr lm y ~ poly(x, 3, raw = TRUE) y ~ poly(x, 3, raw = TRUE) 1
#> npcx y npcy PANEL group label orientation
#> 1 NA 1 NA 1 -1 italic(R)^2~`=`~"0.96" x
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
output.type = "numeric")
#> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (head):"
#> eq.label
#> 1 italic(y)~`=`~-0.00450 + 0.00109*~italic(x) - 2.14%*% 10^{-05}*~italic(x)^2 + 1.06%*% 10^{-06}*~italic(x)^3
#> rr.label adj.rr.label rr.confint.label
#> 1 italic(R)^2~`=`~"0.96" italic(R)[adj]^2~`=`~"0.96" "95% CI [0.95, 0.97]"
#> AIC.label BIC.label
#> 1 plain(AIC)~`=`~"-291.9" plain(BIC)~`=`~"-278.8"
#> f.value.label p.value.label n.label
#> 1 italic(F)[3*","*96]~`=`~"810." italic(P)~`<`~"0.001" italic(n)~`=`~100
#> knots.label grp.label method.label
#> 1 <NA> -1 "method: lm:qr"
#> coefs
#> 1 -4.495631e-03, 1.089644e-03, -2.139937e-05, 1.055387e-06
#> coefs.names
#> 1 (Intercept), poly(x, 3, raw = TRUE)1, poly(x, 3, raw = TRUE)2, poly(x, 3, raw = TRUE)3
#> rr.confint.level rr.confint.low rr.confint.high f.value f.df1 f.df2 r.squared
#> 1 0.95 0.9464423 0.9696371 810.484 3 96 0.9620171
#> adj.r.squared p.value AIC BIC n knots knots.se knots.names
#> 1 0.9608301 5.110349e-68 -291.8639 -278.838 100 NA NA NA
#> fm.method fm.class fm.formula fm.formula.chr x
#> 1 lm:qr lm y ~ poly(x, 3, raw = TRUE) y ~ poly(x, 3, raw = TRUE) 1
#> npcx y npcy PANEL group label orientation
#> 1 NA 1 NA 1 -1 italic(R)^2~`=`~"0.96" x
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
output.type = "numeric")
 #> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (head):"
#> rr.label
#> 1
#> coef.ls
#> 1 -4.495631e-03, 1.089644e-03, -2.139937e-05, 1.055387e-06, 2.268244e-02, 1.935252e-03, 4.440449e-05, 2.890821e-07, -1.981987e-01, 5.630500e-01, -4.819189e-01, 3.650820e+00, 8.433087e-01, 5.747135e-01, 6.309604e-01, 4.255076e-04
#> b_0.constant b_0 b_1 b_2 b_3
#> 1 FALSE -0.004495631 0.001089644 -2.139937e-05 1.055387e-06
#> coefs
#> 1 -4.495631e-03, 1.089644e-03, -2.139937e-05, 1.055387e-06
#> coefs.names
#> 1 (Intercept), poly(x, 3, raw = TRUE)1, poly(x, 3, raw = TRUE)2, poly(x, 3, raw = TRUE)3
#> rr.confint.level rr.confint.low rr.confint.high f.value f.df1 f.df2 r.squared
#> 1 0.95 0.9464423 0.9696371 810.484 3 96 0.9620171
#> adj.r.squared p.value AIC BIC n knots knots.se knots.names
#> 1 0.9608301 5.110349e-68 -291.8639 -278.838 100 NA NA NA
#> fm.method fm.class fm.formula fm.formula.chr x
#> 1 lm:qr lm y ~ poly(x, 3, raw = TRUE) y ~ poly(x, 3, raw = TRUE) 1
#> npcx y npcy PANEL group label orientation
#> 1 NA 1 NA 1 -1 x
# names of the variables
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
dbgfun.data = colnames)
#> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (head):"
#> rr.label
#> 1
#> coef.ls
#> 1 -4.495631e-03, 1.089644e-03, -2.139937e-05, 1.055387e-06, 2.268244e-02, 1.935252e-03, 4.440449e-05, 2.890821e-07, -1.981987e-01, 5.630500e-01, -4.819189e-01, 3.650820e+00, 8.433087e-01, 5.747135e-01, 6.309604e-01, 4.255076e-04
#> b_0.constant b_0 b_1 b_2 b_3
#> 1 FALSE -0.004495631 0.001089644 -2.139937e-05 1.055387e-06
#> coefs
#> 1 -4.495631e-03, 1.089644e-03, -2.139937e-05, 1.055387e-06
#> coefs.names
#> 1 (Intercept), poly(x, 3, raw = TRUE)1, poly(x, 3, raw = TRUE)2, poly(x, 3, raw = TRUE)3
#> rr.confint.level rr.confint.low rr.confint.high f.value f.df1 f.df2 r.squared
#> 1 0.95 0.9464423 0.9696371 810.484 3 96 0.9620171
#> adj.r.squared p.value AIC BIC n knots knots.se knots.names
#> 1 0.9608301 5.110349e-68 -291.8639 -278.838 100 NA NA NA
#> fm.method fm.class fm.formula fm.formula.chr x
#> 1 lm:qr lm y ~ poly(x, 3, raw = TRUE) y ~ poly(x, 3, raw = TRUE) 1
#> npcx y npcy PANEL group label orientation
#> 1 NA 1 NA 1 -1 x
# names of the variables
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
dbgfun.data = colnames)
 #> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (anonymous function):"
#> [1] "eq.label" "rr.label" "adj.rr.label" "rr.confint.label"
#> [5] "AIC.label" "BIC.label" "f.value.label" "p.value.label"
#> [9] "n.label" "knots.label" "grp.label" "method.label"
#> [13] "coefs" "coefs.names" "rr.confint.level" "rr.confint.low"
#> [17] "rr.confint.high" "f.value" "f.df1" "f.df2"
#> [21] "r.squared" "adj.r.squared" "p.value" "AIC"
#> [25] "BIC" "n" "knots" "knots.se"
#> [29] "knots.names" "fm.method" "fm.class" "fm.formula"
#> [33] "fm.formula.chr" "x" "npcx" "y"
#> [37] "npcy" "PANEL" "group" "label"
#> [41] "orientation"
# only data$eq.label
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
output.type = "expression",
dbgfun.data = function(x) {x[["eq.label"]]})
#> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (anonymous function):"
#> [1] "eq.label" "rr.label" "adj.rr.label" "rr.confint.label"
#> [5] "AIC.label" "BIC.label" "f.value.label" "p.value.label"
#> [9] "n.label" "knots.label" "grp.label" "method.label"
#> [13] "coefs" "coefs.names" "rr.confint.level" "rr.confint.low"
#> [17] "rr.confint.high" "f.value" "f.df1" "f.df2"
#> [21] "r.squared" "adj.r.squared" "p.value" "AIC"
#> [25] "BIC" "n" "knots" "knots.se"
#> [29] "knots.names" "fm.method" "fm.class" "fm.formula"
#> [33] "fm.formula.chr" "x" "npcx" "y"
#> [37] "npcy" "PANEL" "group" "label"
#> [41] "orientation"
# only data$eq.label
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
output.type = "expression",
dbgfun.data = function(x) {x[["eq.label"]]})
 #> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (anonymous function):"
#> [1] "italic(y)~`=`~-0.00450 + 0.00109*~italic(x) - 2.14%*% 10^{-05}*~italic(x)^2 + 1.06%*% 10^{-06}*~italic(x)^3"
# only data$eq.label
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
output.type = "text",
dbgfun.data = function(x) {x[["eq.label"]]})
#> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (anonymous function):"
#> [1] "italic(y)~`=`~-0.00450 + 0.00109*~italic(x) - 2.14%*% 10^{-05}*~italic(x)^2 + 1.06%*% 10^{-06}*~italic(x)^3"
# only data$eq.label
if (gginnards.installed)
ggplot(my.data, aes(x, y)) +
geom_point() +
stat_poly_line(formula = formula) +
stat_poly_eq(formula = formula,
geom = "debug_group",
output.type = "text",
dbgfun.data = function(x) {x[["eq.label"]]})
 #> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (anonymous function):"
#> [1] "y = -0.00450 + 0.00109 x - 2.14 10^-05 x^2 + 1.06 10^-06 x^3"
#> [1] "PANEL 1; group(s) -1; 'draw_function()' input 'data' (anonymous function):"
#> [1] "y = -0.00450 + 0.00109 x - 2.14 10^-05 x^2 + 1.06 10^-06 x^3"